AFNI program: afni
Output of -help
**** At the bottom of this Web page are some slide images to
outline the usage of the AFNI Graphical User Interface (GUI).
----------------------------------------------------------------
USAGE 1: read in sessions of 3D datasets (created by to3d, etc.)
----------------------------------------------------------------
afni [options] [session_directory ...]
-bysub This new [01 Feb 2018] option allows you to have 'sessions'
*OR* made up from files scattered across multiple directories.
-BIDS The purpose of this option is to gather all the datasets
corresponding to a single subject identifier, as is done
in the BIDS file hierarchy -- http://bids.neuroimaging.io/
**** There are two methods for using this option.
method (1) ** In the first method, you put one or more subject identifiers,
[OLDER] which are of the form 'sub-XXX' where 'XXX' is some
subject code (it does not have to be exactly 3 characters).
++ If an identifier does NOT start with 'sub-', then that
4 letter string will be added to the front. This allows
you to specify your subjects by their numbers 'XXX' alone.
method (2) ** In the second method, you put one or more directory names,
[NEWER] and all immediate sub-directories whose name starts with
'sub-' will be included. With this method, you can end up
reading in an entire BIDS hierarchy of datasets, which
might take a significant amount of time if there are many
subjects.
**** Note that if an identifier following '-bysub' on the
command line is a directory name that starts with 'sub-',
it will be treated using method (1), not using method (2).
both methods ** In either method, the list of names following '-bysub' ends
with any argument that starts with '-' (or with the end of
all command line arguments).
method (2) ** Each directory on the command line (after all options, and
including any directories directly after the '-bysub' option)
will be scanned recursively (down the file tree) for
subdirectories whose name matches each 'sub-XXX' identifier
exactly. All such subdirectories will have all their
datasets read in (recursively down the file tree) and
put into a single session for viewing in AFNI.
++ In addition, all datasets from all subjects will be
available in the 'All_Datasets' session in the GUI.
(Unless environment variable AFNI_ALL_DATASETS is set to NO)
++ If you do NOT put any directories or subject identifiers
directly after the '-bysub' (or '-BIDS') option, the
program will act as if you put '.' there, and it will
search below the current working directory - the directory
you were 'in' when you started the AFNI GUI.
method (1) ** If a directory on the command line after this option does
NOT have any subdirectories that match any of the '-bysub'
identifiers, then that directory will be read in the normal
way, with all the datasets in that particular directory
(but not subdirectories) read into the session.
both methods ** Please note that '-bysub' sessions will NOT be rescanned
for new datasets that might get placed there after the
AFNI GUI starts, unlike normal (single directory) sessions.
method (1) ** Example (method 1):
afni -bysub 10506 50073 - ~/data/OpenFMRI/ds000030
This will open the data for subjects 10506 and 50073 from
the data at the specified directory -- presumably the
data downloaded from https://openfmri.org/dataset/ds000030/
++ If directory sub-10506 is found and has (say) sub-directories
anat beh dwi func
all AFNI-readable datasets from these sub-directories will
be input and collected into one session, to be easily
viewed together.
++ Because of the recursive search, if a directory named (e.g.)
derivatives/sub-10506
is found underneath ~/data/OpenFMRI/ds000030, all the
datasets found underneath that will also be put into the
same session, so they can be viewed with the 'raw' data.
++ In this context, 'dataset' also means .png and .jpg files
found in the sub-XXX directories. These images can be
opened in the AFNI GUI using the Axial image viewer.
(You might want to turn the AFNI crosshairs off!)
++++ If you do want .png and .jpg files read into AFNI,
set environment variable AFNI_IMAGE_DATASETS to 'Y'.
++ You can put multiple subject IDs after '-bysub', as
in the example above. You can also use the '-bysub' option
more than once, if you like. Each distinct subect ID will
get a distinct AFNI session in the GUI.
method (2) ** Example (method 2):
afni -bysub ~/data/OpenFMRI/ds000030
This will read in all datasets from all subjects. In this
particular example, there are hundreds of subjects, so this
command may not actually be a good idea - unless you want to
go get a cup of chai or coffee, and then sip it very slowly.
** Example (method 2):
afni -BIDS
This will read all 'sub-*' directories from the current
working directory, and is the same as 'afni -BIDS .'
As noted earlier, this recursive operation may take a long
time (especially if the datasets are compressed), as AFNI
reads the headers from ALL datasets as it finds them,
to build a table for you to use in the 'OverLay' and
'UnderLay' dataset choosers.
-all_dsets Read in all datasets from all listed folders together.
Has the same effect as choosing 'All_Datasets' in the GUI.
Example: afni -all_dsets dir1 dir2 dir3
Can be set to default in .afnirc with ALL_DSETS_STARTUP = YES.
Overridden silently by AFNI_ALL_DATASETS = NO.
-purge Conserve memory by purging unused datasets from memory.
[Use this if you run out of memory when running AFNI.]
[This will slow the code down, so use only if needed.]
[When a dataset is needed, it will be re-read from disk.]
-posfunc Start up the color 'pbar' to use only positive function values.
-R Recursively search each session_directory for more session
subdirectories.
WARNING: This will descend the entire filesystem hierarchy from
each session_directory given on the command line. On a
large disk, this may take a long time. To limit the
recursion to 5 levels (for example), use -R5.
** Use of '-bysub' disables recursive descent, since '-bysub'
will do that for you.
-no1D Tells AFNI not to read *.1D timeseries files from
the dataset directories. The *.1D files in the
directories listed in the AFNI_TSPATH environment
variable will still be read (if this variable is
not set, then './' will be scanned for *.1D files).
-nocsv Each of these option flags does the same thing (i.e.,
-notsv they are synonyms): each tells AFNI not to read
-notcsv *.csv or *.tsv files from the dataset directories.
You can also set env AFNI_SKIP_TCSV_SCAN = YES to the
same effect.
-unique Tells the program to create a unique set of colors
for each AFNI controller window. This allows
different datasets to be viewed with different
grayscales or colorscales. Note that -unique
will only work on displays that support 12 bit
PseudoColor (e.g., SGI workstations) or TrueColor.
-orient code Tells afni the orientation in which to display
x-y-z coordinates (upper left of control window).
The code must be 3 letters, one each from the
pairs {R,L} {A,P} {I,S}. The first letter gives
the orientation of the x-axis, the second the
orientation of the y-axis, the third the z-axis:
R = right-to-left L = left-to-right
A = anterior-to-posterior P = posterior-to-anterior
I = inferior-to-superior S = superior-to-inferior
The default code is RAI ==> DICOM order. This can
be set with the environment variable AFNI_ORIENT.
As a special case, using the code 'flipped' is
equivalent to 'LPI' (this is for Steve Rao).
-noplugins Tells the program not to load plugins.
(Plugins can also be disabled by setting the
environment variable AFNI_NOPLUGINS.)
-seehidden Tells the program to show you which plugins
are hidden.
-DAFNI_ALLOW_ALL_PLUGINS=YES
Tells the program NOT to hide plugins from you.
Note that there are a lot of hidden plugins,
most of which are not very useful!
-yesplugouts Tells the program to listen for plugouts.
(Plugouts can also be enabled by setting the
environment variable AFNI_YESPLUGOUTS.)
-YESplugouts Makes the plugout code print out lots of messages
(useful for debugging a new plugout).
-noplugouts Tells the program NOT to listen for plugouts.
(This option is available to override
the AFNI_YESPLUGOUTS environment variable.)
-skip_afnirc Tells the program NOT to read the file .afnirc
in the home directory. See README.setup for
details on the use of .afnirc for initialization.
-layout fn Tells AFNI to read the initial windows layout from
file 'fn'. If this option is not given, then
environment variable AFNI_LAYOUT_FILE is used.
If neither is present, then AFNI will do whatever
it feels like.
-niml If present, turns on listening for NIML-formatted
data from SUMA. Can also be turned on by setting
environment variable AFNI_NIML_START to YES.
-np PORT_OFFSET: Provide a port offset to allow multiple instances of
AFNI <--> SUMA, AFNI <--> 3dGroupIncorr, or any other
programs that communicate together to operate on the same
machine.
All ports are assigned numbers relative to PORT_OFFSET.
The same PORT_OFFSET value must be used on all programs
that are to talk together. PORT_OFFSET is an integer in
the inclusive range [1025 to 65500].
When you want to use multiple instances of communicating programs,
be sure the PORT_OFFSETS you use differ by about 50 or you may
still have port conflicts. A BETTER approach is to use -npb below.
-npq PORT_OFFSET: Like -np, but more quiet in the face of adversity.
-npb PORT_OFFSET_BLOC: Similar to -np, except it is easier to use.
PORT_OFFSET_BLOC is an integer between 0 and
MAX_BLOC. MAX_BLOC is around 4000 for now, but
it might decrease as we use up more ports in AFNI.
You should be safe for the next 10 years if you
stay under 2000.
Using this function reduces your chances of causing
port conflicts.
See also afni and suma options: -list_ports and -port_number for
information about port number assignments.
You can also provide a port offset with the environment variable
AFNI_PORT_OFFSET. Using -np overrides AFNI_PORT_OFFSET.
-max_port_bloc: Print the current value of MAX_BLOC and exit.
Remember this value can get smaller with future releases.
Stay under 2000.
-max_port_bloc_quiet: Spit MAX_BLOC value only and exit.
-num_assigned_ports: Print the number of assigned ports used by AFNI
then quit.
-num_assigned_ports_quiet: Do it quietly.
Port Handling Examples:
-----------------------
Say you want to run three instances of AFNI <--> SUMA.
For the first you just do:
suma -niml -spec ... -sv ... &
afni -niml &
Then for the second instance pick an offset bloc, say 1 and run
suma -niml -npb 1 -spec ... -sv ... &
afni -niml -npb 1 &
And for yet another instance:
suma -niml -npb 2 -spec ... -sv ... &
afni -niml -npb 2 &
etc.
Since you can launch many instances of communicating programs now,
you need to know wich SUMA window, say, is talking to which AFNI.
To sort this out, the titlebars now show the number of the bloc
of ports they are using. When the bloc is set either via
environment variables AFNI_PORT_OFFSET or AFNI_PORT_BLOC, or
with one of the -np* options, window title bars change from
[A] to [A#] with # being the resultant bloc number.
In the examples above, both AFNI and SUMA windows will show [A2]
when -npb is 2.
-list_ports List all port assignments and quit
-port_number PORT_NAME: Give port number for PORT_NAME and quit
-port_number_quiet PORT_NAME: Same as -port_number but writes out
number only
-available_npb: Find the first available block of port numbers,
print it to stdout and quit
The value can be used to set the -npb option for
a new set of chatty AFNI/SUMA/etc group.
-available_npb_quiet: Just print the block number to stdout and quit.
-com ccc This option lets you specify 'command strings' to
drive AFNI after the program startup is completed.
Legal command strings are described in the file
README.driver. More than one '-com' option can
be used, and the commands will be executed in
the order they are given on the command line.
N.B.: Most commands to AFNI contain spaces, so the 'ccc'
command strings will need to be enclosed in quotes.
-comsep 'c' Use character 'c' as a separator for commands.
In this way, you can put multiple commands in
a single '-com' option. Default separator is ';'.
N.B.: The command separator CANNOT be alphabetic or
numeric (a..z, A..Z, 0..9) or whitespace or a quote!
N.B.: -comsep should come BEFORE any -com option that
uses a non-semicolon separator!
Example: -com 'OPEN_WINDOW axialimage; SAVE_JPEG axialimage zork; QUIT'
N.B.: You can also put startup commands (one per line) in
the file '~/.afni.startup_script'. For example,
OPEN_WINDOW axialimage
to always open the axial image window on startup.
* If no session_directories are given, then the program will use
the current working directory (i.e., './').
* The maximum number of sessions is now set to 199.
* The maximum number of datasets per session is 8192.
* To change these maximums, you must edit file '3ddata.h' and then
recompile this program.
Global Options (available to all AFNI/SUMA programs)
-h: Mini help, at time, same as -help in many cases.
-help: The entire help output
-HELP: Extreme help, same as -help in majority of cases.
-h_view: Open help in text editor. AFNI will try to find a GUI editor
-hview : on your machine. You can control which it should use by
setting environment variable AFNI_GUI_EDITOR.
-h_web: Open help in web browser. AFNI will try to find a browser.
-hweb : on your machine. You can control which it should use by
setting environment variable AFNI_GUI_EDITOR.
-h_find WORD: Look for lines in this programs's -help output that match
(approximately) WORD.
-h_raw: Help string unedited
-h_spx: Help string in sphinx loveliness, but do not try to autoformat
-h_aspx: Help string in sphinx with autoformatting of options, etc.
-all_opts: Try to identify all options for the program from the
output of its -help option. Some options might be missed
and others misidentified. Use this output for hints only.
-overwrite: Overwrite existing output dataset.
Equivalent to setting env. AFNI_DECONFLICT=OVERWRITE
-ok_1D_text: Zero out uncommented text in 1D file.
Equivalent to setting env. AFNI_1D_ZERO_TEXT=YES
-Dname=val: Set environment variable 'name' to value 'val'
For example: -DAFNI_1D_ZERO_TEXT=YES
-Vname=: Print value of environment variable 'name' to stdout and quit.
This is more reliable that the shell's env query because it would
include envs set in .afnirc files and .sumarc files for SUMA
programs.
For example: -VAFNI_1D_ZERO_TEXT=
-skip_afnirc: Do not read the afni resource (like ~/.afnirc) file.
-pad_to_node NODE: Output a full dset from node 0 to MAX_NODE-1
** Instead of directly setting NODE to an integer you
can set NODE to something like:
ld120 (or rd17) which sets NODE to be the maximum
node index on an Icosahedron with -ld 120. See
CreateIcosahedron for details.
d:DSET.niml.dset which sets NODE to the maximum node found
in dataset DSET.niml.dset.
** This option is for surface-based datasets only.
Some programs may not heed it, so check the output if
you are not sure.
-pif SOMETHING: Does absolutely nothing but provide for a convenient
way to tag a process and find it in the output of ps -a
-echo_edu: Echos the entire command line to stdout (without -echo_edu)
for edification purposes
SPECIAL PURPOSE ARGUMENTS TO ADD *MORE* ARGUMENTS TO THE COMMAND LINE
------------------------------------------------------------------------
Arguments of the following form can be used to create MORE command
line arguments -- the principal reason for using these type of arguments
is to create program command lines that are beyond the limit of
practicable scripting. (For one thing, Unix command lines have an
upper limit on their length.) This type of expanding argument makes
it possible to input thousands of files into an AFNI program command line.
The generic form of these arguments is (quotes, 'single' or "double",
are required for this type of argument):
'<<XY list'
where X = I for Include (include strings from file)
or X = G for Glob (wildcard expansion)
where Y = M for Multi-string (create multiple arguments from multiple strings)
or Y = 1 for One-string (all strings created are put into one argument)
Following the XY modifiers, a list of strings is given, separated by spaces.
* For X=I, each string in the list is a filename to be read in and
included on the command line.
* For X=G, each string in the list is a Unix style filename wildcard
expression to be expanded and the resulting filenames included
on the command line.
In each case, the '<<XY list' command line argument will be removed and
replaced by the results of the expansion.
* '<<GM wildcards'
Each wildcard string will be 'globbed' -- expanded from the names of
files -- and the list of files found this way will be stored in a
sequence of new arguments that replace this argument:
'<<GM ~/Alice/*.nii ~/Bob/*.nii'
might expand into a list of hundreds of separate datasets.
* Why use this instead of just putting the wildcards on the command
line? Mostly to get around limits on the length of Unix command lines.
* '<<G1 wildcards'
The difference from the above case is that after the wildcard expansion
strings are found, they are catenated with separating spaces into one
big string. The only use for this in AFNI is for auto-catenation of
multiple datasets into one big dataset.
* '<<IM filenames'
Each filename string will result in the contents of that text file being
read in, broken at whitespace into separate strings, and the resulting
collection of strings will be stored in a sequence of new arguments
that replace this argument. This type of argument can be used to input
large numbers of files which are listed in an external file:
'<<IM Bob.list.txt'
which could in principle result in reading in thousands of datasets
(if you've got the RAM).
* This type of argument is in essence an internal form of doing something
like `cat filename` using the back-quote shell operator on the command
line. The only reason this argument (or the others) was implemented is
to get around the length limits on the Unix command line.
* '<<I1 filenames'
The difference from the above case is that after the files are read
and their strings are found, they are catenated with separating spaces
into one big string. The only use for this in AFNI is for auto-catenation
of multiple datasets into one big dataset.
* 'G', 'M', and 'I' can be lower case, as in '<<gm'.
* 'glob' is Unix jargon for wildcard expansion:
https://en.wikipedia.org/wiki/Glob_(programming)
* If you set environment variable AFNI_GLOB_SELECTORS to YES,
then the wildcard expansion with '<<g' will not use the '[...]'
construction as a Unix wildcard. Instead, it will expand the rest
of the wildcard and then append the '[...]' to the results:
'<<gm fred/*.nii[1..100]'
would expand to something like
fred/A.nii[1..100] fred/B.nii[1..100] fred/C.nii[1..100]
This technique is a way to preserve AFNI-style sub-brick selectors
and have them apply to a lot of files at once.
Another example:
3dttest++ -DAFNI_GLOB_SELECTORS=YES -brickwise -prefix Junk.nii \
-setA '<<gm sub-*/func/*rest_bold.nii.gz[0..100]'
* However, if you want to put sub-brick selectors on the '<<im' type
of input, you will have to do that in the input text file itself
(for each input filename in that file).
* BE CAREFUL OUT THERE!
------------------------------------------------------------------------
-------------------------------------------------------
USAGE 2: read in datasets specified on the command line
-------------------------------------------------------
afni -dset [options] dname1 dname2 ...
where 'dname1' is the name of a dataset, etc. With this option, only
the chosen datasets are read in, and they are all put in the same
'session'. Follower datasets are not created.
* If you wish to be very tricksy, you can read in .1D files as datasets
using the \' transpose syntax, as in
afni Fred.1D\'
However, this isn't very useful (IMHO).
* AFNI can also read image files (.jpg and .png) from the command line.
For just viewing images, the 'aiv' program (AFNI image viewer) is
simpler; but unlike aiv, you can do basic image processing on an
image 'dataset' using the AFNI GUI's feature. Sample command:
afni *.jpg
Each image file is a single 'dataset'; to switch between images,
use the 'Underlay' button. To view an image, open the 'Axial' viewer.
INPUT DATASET NAMES
-------------------
An input dataset is specified using one of these forms:
'prefix+view', 'prefix+view.HEAD', or 'prefix+view.BRIK'.
You can also add a sub-brick selection list after the end of the
dataset name. This allows only a subset of the sub-bricks to be
read in (by default, all of a dataset's sub-bricks are input).
A sub-brick selection list looks like one of the following forms:
fred+orig[5] ==> use only sub-brick #5
fred+orig[5,9,17] ==> use #5, #9, and #17
fred+orig[5..8] or [5-8] ==> use #5, #6, #7, and #8
fred+orig[5..13(2)] or [5-13(2)] ==> use #5, #7, #9, #11, and #13
Sub-brick indexes start at 0. You can use the character '$'
to indicate the last sub-brick in a dataset; for example, you
can select every third sub-brick by using the selection list
fred+orig[0..$(3)]
N.B.: The sub-bricks are read in the order specified, which may
not be the order in the original dataset. For example, using
fred+orig[0..$(2),1..$(2)]
will cause the sub-bricks in fred+orig to be input into memory
in an interleaved fashion. Using
fred+orig[$..0]
will reverse the order of the sub-bricks.
N.B.: You may also use the syntax <a..b> after the name of an input
dataset to restrict the range of values read in to the numerical
values in a..b, inclusive. For example,
fred+orig[5..7]<100..200>
creates a 3 sub-brick dataset with values less than 100 or
greater than 200 from the original set to zero.
If you use the <> sub-range selection without the [] sub-brick
selection, it is the same as if you had put [0..$] in front of
the sub-range selection.
N.B.: Datasets using sub-brick/sub-range selectors are treated as:
- 3D+time if the dataset is 3D+time and more than 1 brick is chosen
- otherwise, as bucket datasets (-abuc or -fbuc)
(in particular, fico, fitt, etc datasets are converted to fbuc!)
N.B.: The characters '$ ( ) [ ] < >' are special to the shell,
so you will have to escape them. This is most easily done by
putting the entire dataset plus selection list inside forward
single quotes, as in 'fred+orig[5..7,9]', or double quotes "x".
CATENATED AND WILDCARD DATASET NAMES
------------------------------------
Datasets may also be catenated or combined in memory, as if one first
ran 3dTcat or 3dbucket.
An input with space-separated elements will be read as a concatenated
dataset, as with 'dset1+tlrc dset2+tlrc dset3+tlrc', or with paths,
'dir/dset1+tlrc dir/dset2+tlrc dir/dset3+tlrc'.
The datasets will be combined (as if by 3dTcat) and then treated as a
single input dataset. Note that the quotes are required to specify
them as a single argument.
Sub-brick selection using '[]' works with space separated dataset
names. If the selector is at the end, it is considered global and
applies to all inputs. Otherwise, it applies to the adjacent input.
For example:
local: 'dset1+tlrc[2,3] dset2+tlrc[7,0,1] dset3+tlrc[5,0,$]'
global: 'dset1+tlrc dset2+tlrc dset3+tlrc[5,6]'
N.B. If AFNI_PATH_SPACES_OK is set to Yes, will be considered as part
of the dataset name, and not as a separator between them.
Similar treatment applies when specifying datasets using a wildcard
pattern, using '*' or '?', as in: 'dset*+tlrc.HEAD'. Any sub-brick
selectors would apply to all matching datasets, as with:
'dset*+tlrc.HEAD[2,5,3]'
N.B.: complete filenames are required when using wildcard matching,
or no files will exist to match, e.g. 'dset*+tlrc' would not work.
N.B.: '[]' are processed as sub-brick or time point selectors. They
are therefore not allowed as wildcard characters in this context.
Space and wildcard catenation can be put together. In such a case,
spaces divide the input into wildcard pieces, which are processed
individually.
Examples (each is processed as a single, combined dataset):
'dset1+tlrc dset2+tlrc dset3+tlrc'
'dset1+tlrc dset2+tlrc dset3+tlrc[2,5,3]'
'dset1+tlrc[3] dset2+tlrc[0,1] dset3+tlrc[3,0,1]'
'dset*+tlrc.HEAD'
'dset*+tlrc.HEAD[2,5,3]'
'dset1*+tlrc.HEAD[0,1] dset2*+tlrc.HEAD[7,8]'
'group.*/subj.*/stats*+tlrc.HEAD[7]'
CALCULATED DATASETS
-------------------
Datasets may also be specified as runtime-generated results from
program 3dcalc. This type of dataset specifier is enclosed in
quotes, and starts with the string '3dcalc(':
'3dcalc( opt opt ... opt )'
where each 'opt' is an option to program 3dcalc; this program
is run to generate a dataset in the directory given by environment
variable TMPDIR (default=/tmp). This dataset is then read into
memory, locked in place, and deleted from disk. For example
afni -dset '3dcalc( -a r1+orig -b r2+orig -expr 0.5*(a+b) )'
will let you look at the average of datasets r1+orig and r2+orig.
N.B.: using this dataset input method will use lots of memory!
-------------------------------
GENERAL OPTIONS (for any usage)
-------------------------------
-papers Prints out the list of AFNI papers, and exits.
-q Tells afni to be 'quiet' on startup
-Dname=val Sets environment variable 'name' to 'val' inside AFNI;
will supersede any value set in .afnirc.
-gamma gg Tells afni that the gamma correction factor for the
monitor is 'gg' (default gg is 1.0; greater than
1.0 makes the image contrast larger -- this may
also be adjusted interactively)
-install Tells afni to install a new X11 Colormap. This only
means something for PseudoColor displays. Also, it
usually cause the notorious 'technicolor' effect.
-ncolors nn Tells afni to use 'nn' gray levels for the image
displays (default is 80)
-xtwarns Tells afni to show any Xt warning messages that may
occur; the default is to suppress these messages.
-XTWARNS Trigger a debug trace when an Xt warning happens.
-tbar name Uses 'name' instead of 'AFNI' in window titlebars.
-flipim and The '-flipim' option tells afni to display images in the
-noflipim 'flipped' radiology convention (left on the right).
The '-noflipim' option tells afni to display left on
the left, as neuroscientists generally prefer. This
latter mode can also be set by the Unix environment
variable 'AFNI_LEFT_IS_LEFT'. The '-flipim' mode is
the default.
-trace Turns routine call tracing on, for debugging purposes.
-TRACE Turns even more verbose tracing on, for more debugging.
-motif_ver Show the applied motif version string.
-no_detach Do not detach from the terminal.
-no_frivolities Turn of all frivolities/fun stuff.
-get_processed_env Show applied AFNI/NIFTI environment variables.
-global_opts Show options that are global to all AFNI programs.
-goodbye [n] Print a 'goodbye' message and exit (just for fun).
If an integer is supplied afterwards, will print that
many (random) goodbye messages.
-startup [n] Similar to '-goodbye', but for startup tips.
[If you want REAL fun, use '-startup ALL'.]
-julian Print out the current Julian date and exit.
-ver Print the current AFNI version and compile date, then exit.
Useful to check how up-to-date you are (or aren't).
-vnum Print just the current AFNI version number (i.e.,
AFNI_A.B.C), then exit.
-package Print just the current AFNI package (i.e.,
linux_ubuntu_12_64, macos_10.12_local, etc.),
then exit.
-tips Print the tips for the GUI, such as key presses
and other useful advice. This is the same file that
would be displayed with the 'AFNI Tips' button in the
GUI controller. Exit after display.
-env Print the environment variables for AFNI, which a user
might set in their ~/.afnirc file (wait, you *do*
have one on your computer, right?).
Exit after display.
-show show the package name
-x_have_MACOS_FORCE_EXPOSE
show whether compiled with MACOS_FORCE_EXPOSE
-x_needsX11Redraw
show whether resize events will cause redraws
N.B.: Many of these options, as well as the initial color set up,
can be controlled by appropriate X11 resources. See the
files AFNI.Xdefaults and README.environment for instructions
and examples.
-----------------------------------------------------------
Options that affect X11 Display properties: '-XXXsomething'
-----------------------------------------------------------
My intent with these options is that you use them in aliases
or shell scripts, to let you setup specific appearances for
multiple copies of AFNI. For example, put the following
command in your shell startup file (e.g., ~/.cshrc or ~/.bashrc)
alias ablue afni -XXXfgcolor white -XXXbgcolor navyblue
Then the command 'ablue' will start AFNI with a blue background
and using white for the default text color.
Note that these options set 'properties' on the X11 server,
which might survive after AFNI exits (especially if AFNI crashes).
If for some reason these settings cause trouble after AFNI
exits, use the option '-XXX defaults' to reset the X11
properties for AFNI back to their default values.
Also note that each option is of the form '-XXXsomething', followed
by a single argument.
-XXXfgcolor colorname = set the 'foreground' color (text color)
to 'colorname'
[default = yellow]
++ This should be a bright color, to contrast
the background color.
++ You can find a list of X11 color names at
https://en.wikipedia.org/wiki/X11_color_names
However, if you use a name like Dark Cyan
(with a space inside the name), you must
put the name in quotes: 'Dark Cyan', or remove
the space: DarkCyan.
++ Another way to specify X11 colors is in hexadecimal,
as in '#rgb' or '#rrggbb', where the letters shown
are replaced by hex values from 0 to f. For example,
'#ffcc00' is an orange-yellow mixture.
-XXXbgcolor colorname = set the 'background' color to 'colorname'
[default = gray22]
++ This should be a somewhat dark color,
or parts of the interface may be hard
to read.
++ EXAMPLE:
afni -XXXfgcolor #00ffaa -XXXbgcolor #330000 -plus
You can create command aliases to open AFNI with
different color schemes, to make your life simpler.
-XXXfontsize plus = set all the X11 fonts used by AFNI to be one
*OR* size larger ('plus') or to be one size smaller
-XXXfontsize minus ('minus'). The 'plus' version I find useful for
*OR* a screen resolution of about 100 dots per inch
-XXXfontsize big (40 dots per cm) -- you can find what the system
*OR* thinks your screen resolution is by the command
-big xdpyinfo | grep -i resolution
*OR* ++ Applying 'plus' twice is the same as 'big'.
-plus ++ Using 'big' will use large Adobe Courier fonts.
*OR* ++ Alternatively, you can control each of the 4 fonts
-minus that AFNI uses, via the 4 following options ...
*OR* ++ You can also set the fontsize for your copy
-norm of AFNI in your ~/.afnirc file by setting
environment variable AFNI_FONTSIZE to one of:
big *OR* minus *or* plus
++ Using 'norm' gives the default AFNI font sizes.
-XXXfontA fontname = set the X11 font name for the main AFNI
controller
[default = 9x15bold]
++ To see a list of all X11 font names, type the command
xlsfonts | more
*or* more elaborately (to show only fixed width fonts):
xlsfonts | grep -e '-[cm]-' | grep -e '-iso8859-1$' | grep -e '-medium-' \
| grep -e '-r-normal-' | grep -v -e '-0-0-' | sort -t '-' -k 8 -n | uniq
++ It is best to use a fixed width font
(e.g., not Helvetica), or the AFNI buttons
won't line up nicely!
++ If you use an illegal font name here, you
might make it hard to use the AFNI GUI!
++ The default fonts are chosen for 'normal' screen
resolutions (about 72 dots per inch = 28 dots per cm).
For higher resolutions ('Retina'), you might
want to use larger fonts. Adding these
'-XXXfont?' options is one way to address this
problem. (Also see '-plus' above.)
++ An example of two quite large fonts on my computer
(which at this time has a 108 dot per inch display):
'-adobe-courier-bold-r-normal--34-240-100-100-m-200-iso8859-1
'-b&h-lucidatypewriter-medium-r-normal-sans-34-240-100-100-m-200-iso8859-1'
Note that to use the latter font on the command line,
you have to enclose the name in quotes, as shown above,
since the 'foundry name' includes the character '&'.
To use it in an alias, you need to do something like
alias abig -XXXfontA '-b\&h-lucidatypewriter-medium-r-normal-sans-34-240-100-100-m-200-iso8859-1'
++ When setting the fonts, it is often helpful
to set the colors as well.
-XXXfontB fontname = set the X11 font name for somewhat smaller text
[default = 8x13bold]
-XXXfontC fontname = set the X11 font name for even smaller text
[default = 7x13]
-XXXfontD fontname = set the X11 font name for the smallest text
[default = 6x10]
-XXX defaults = set the X11 properties to the AFNI defaults
(the purpose of this is to restore things )
(to normal if the X11 settings get mangled)
-XXXnpane P = set the number of 'panes' in the continuous
colorscale to the value 'P', where P is an
even integer between 256 and 2048 (inclusive).
Probably will work best if P is an integral
multiple of 256 (e.g., 256, 512, 1024, 2048).
[This option is for the mysterious Dr ZXu.]
--------------------------------------
Educational and Informational Material
--------------------------------------
* The presentations used in our AFNI teaching classes at the NIH can
all be found at
https://afni.nimh.nih.gov/pub/dist/edu/latest/ (PowerPoint directories)
https://afni.nimh.nih.gov/pub/dist/edu/latest/afni_handouts/ (PDF directory)
* And for the interactive AFNI program in particular, see
https://afni.nimh.nih.gov/pub/dist/edu/latest/afni01_intro/afni01_intro.pdf
https://afni.nimh.nih.gov/pub/dist/edu/latest/afni03_interactive/afni03_interactive.pdf
* For the -help on all AFNI programs, plus the README files, and more, please see
https://afni.nimh.nih.gov/pub/dist/doc/program_help/index.html
* For indvidualized help with AFNI problems, and to keep up with AFNI news, please
use the AFNI Message Board:
https://discuss.afni.nimh.nih.gov
* If an AFNI program crashes, please include the EXACT error messages it outputs
in your message board posting, as well as any other information needed to
reproduce the problem. Just saying 'program X crashed, what's the problem?'
is not helpful at all! In all message board postings, detail and context
are highly relevant.
* Also, be sure your AFNI distribution is up-to-date. You can check the date
on your copy with the command 'afni -ver'. If it is more than a few months
old, you should update your AFNI binaries and try the problematic command
again -- it is quite possible the problem you encountered was already fixed!
****************************************************
***** This is a list of papers about AFNI, SUMA, *****
****** and various algorithms implemented therein ******
----------------------------------------------------------------------------
RW Cox.
AFNI: Software for analysis and visualization of functional
magnetic resonance neuroimages. Computers and Biomedical Research,
29: 162-173, 1996.
* The very first AFNI paper, and the one I prefer you cite if you want
to refer to the AFNI package as a whole.
* https://sscc.nimh.nih.gov/sscc/rwcox/papers/CBM_1996.pdf
----------------------------------------------------------------------------
RW Cox, A Jesmanowicz, JS Hyde.
Real-time functional magnetic resonance imaging.
Magnetic Resonance in Medicine, 33: 230-236, 1995.
* The first paper on realtime FMRI; describes the algorithm used in
in the realtime plugin for time series regression analysis.
* https://sscc.nimh.nih.gov/sscc/rwcox/papers/Realtime_FMRI.pdf
----------------------------------------------------------------------------
RW Cox, JS Hyde.
Software tools for analysis and visualization of FMRI Data.
NMR in Biomedicine, 10: 171-178, 1997.
* A second paper about AFNI and design issues for FMRI software tools.
----------------------------------------------------------------------------
RW Cox, A Jesmanowicz.
Real-time 3D image registration for functional MRI.
Magnetic Resonance in Medicine, 42: 1014-1018, 1999.
* Describes the algorithm used for image registration in 3dvolreg
and in the realtime plugin.
* The first paper to demonstrate realtime MRI volume image
registration running on a standard workstation (not a supercomputer).
* https://sscc.nimh.nih.gov/sscc/rwcox/papers/RealtimeRegistration.pdf
----------------------------------------------------------------------------
ZS Saad, KM Ropella, RW Cox, EA DeYoe.
Analysis and use of FMRI response delays.
Human Brain Mapping, 13: 74-93, 2001.
* Describes the algorithm used in 3ddelay (cf. '3ddelay -help').
* https://sscc.nimh.nih.gov/sscc/rwcox/papers/Delays2001.pdf
----------------------------------------------------------------------------
ZS Saad, RC Reynolds, BD Argall, S Japee, RW Cox.
SUMA: An interface for surface-based intra- and inter-subject analysis
within AFNI. 2004 IEEE International Symposium on Biomedical Imaging:
from Nano to Macro. IEEE, Arlington VA, pp. 1510-1513.
* A brief description of SUMA.
* https://dx.doi.org/10.1109/ISBI.2004.1398837
* https://sscc.nimh.nih.gov/sscc/rwcox/papers/SUMA2004paper.pdf
----------------------------------------------------------------------------
ZS Saad, G Chen, RC Reynolds, PP Christidis, KR Hammett, PSF Bellgowan,
RW Cox.
FIAC Analysis According to AFNI and SUMA.
Human Brain Mapping, 27: 417-424, 2006.
* Describes how we used AFNI to analyze the FIAC contest data.
* https://dx.doi.org/10.1002/hbm.20247
* https://sscc.nimh.nih.gov/sscc/rwcox/papers/FIAC_AFNI_2006.pdf
----------------------------------------------------------------------------
BD Argall, ZS Saad, MS Beauchamp.
Simplified intersubject averaging on the cortical surface using SUMA.
Human Brain Mapping 27: 14-27, 2006.
* Describes the 'standard mesh' surface approach used in SUMA.
* https://dx.doi.org/10.1002/hbm.20158
* https://sscc.nimh.nih.gov/sscc/rwcox/papers/SUMA2006paper.pdf
----------------------------------------------------------------------------
ZS Saad, DR Glen, G Chen, MS Beauchamp, R Desai, RW Cox.
A new method for improving functional-to-structural MRI alignment
using local Pearson correlation. NeuroImage 44: 839-848, 2009.
* Describes the algorithm used in 3dAllineate (and thence in
align_epi_anat.py) for EPI-to-structural volume image registration.
* https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2649831/
* https://dx.doi.org/10.1016/j.neuroimage.2008.09.037
* https://sscc.nimh.nih.gov/sscc/rwcox/papers/LocalPearson2009.pdf
----------------------------------------------------------------------------
H Sarin, AS Kanevsky, SH Fung, JA Butman, RW Cox, D Glen, R Reynolds, S Auh.
Metabolically stable bradykinin B2 receptor agonists enhance transvascular
drug delivery into malignant brain tumors by increasing drug half-life.
Journal of Translational Medicine, 7: #33, 2009.
* Describes the method used in AFNI for modeling dynamic contrast enhanced
(DCE) MRI for analysis of brain tumors.
* https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2689161/
* https://dx.doi.org/10.1186/1479-5876-7-33
----------------------------------------------------------------------------
HJ Jo, ZS Saad, WK Simmons, LA Milbury, RW Cox.
Mapping sources of correlation in resting state FMRI, with artifact detection
and removal. NeuroImage, 52: 571-582, 2010.
* Describes the ANATICOR method for de-noising FMRI datasets.
* https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2897154/
* https://dx.doi.org/10.1016/j.neuroimage.2010.04.246
----------------------------------------------------------------------------
A Vovk, RW Cox, J Stare, D Suput, ZS Saad.
Segmentation Priors From Local Image Properties: Without Using Bias Field
Correction, Location-based Templates, or Registration.
Neuroimage, 55: 142-152, 2011.
* Describes the earliest basis for 3dSeg.
* https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3031751/
* https://dx.doi.org/10.1016/j.neuroimage.2010.11.082
----------------------------------------------------------------------------
G Chen, ZS Saad, DR Glen, JP Hamilton, ME Thomason, IH Gotlib, RW Cox.
Vector Autoregression, Structural Equation Modeling, and Their Synthesis in
Neuroimaging Data Analysis.
Computers in Biology and Medicine, 41: 1142-1155, 2011.
* Describes the method implemented in 1dSVAR (Structured Vector AutoRegression).
* https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3223325/
* https://dx.doi.org/10.1016/j.compbiomed.2011.09.004
----------------------------------------------------------------------------
RW Cox.
AFNI: what a long strange trip it's been. NeuroImage, 62: 747-765, 2012.
* A Brief History of AFNI, from its inception to speculation about the future.
* https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3246532/
* https://dx.doi.org/10.1016/j.neuroimage.2011.08.056
----------------------------------------------------------------------------
ZS Saad, RC Reynolds.
SUMA. Neuroimage. 62: 768-773, 2012.
* The biography of SUMA.
* https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3260385/
* https://dx.doi.org/10.1016/j.neuroimage.2011.09.016
----------------------------------------------------------------------------
G Chen, ZS Saad, AR Nath, MS Beauchamp, RW Cox.
FMRI Group Analysis Combining Effect Estimates and Their Variances.
Neuroimage, 60: 747-765, 2012.
* The math behind 3dMEMA (Mixed Effects Meta-Analysis) -- AKA super-3dttest.
* https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3404516/
* https://dx.doi.org/10.1016/j.neuroimage.2011.12.060
----------------------------------------------------------------------------
ZS Saad, SJ Gotts, K Murphy, G Chen, HJ Jo, A Martin, RW Cox.
Trouble at Rest: How Correlation Patterns and Group Differences Become
Distorted After Global Signal Regression.
Brain Connectivity, 2: 25-32, 2012.
* Our first paper on why Global Signal Regression in resting state FMRI is
a bad idea when doing any form of group analysis.
* https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3484684/
* https://dx.doi.org/10.1089/brain.2012.0080
----------------------------------------------------------------------------
SJ Gotts, WK Simmons, LA Milbury, GL Wallace, RW Cox, A Martin.
Fractionation of Social Brain Circuits in Autism Spectrum Disorders.
Brain, 135: 2711-2725, 2012.
* In our humble opinion, this shows how to use resting state FMRI correctly when
making inter-group comparisons (hint: no global signal regression is used).
* https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3437021/
* https://dx.doi.org/10.1093/brain/aws160
----------------------------------------------------------------------------
HJ Jo, ZS Saad, SJ Gotts, A Martin, RW Cox.
Quantifying Agreement between Anatomical and Functional Interhemispheric
Correspondences in the Resting Brain.
PLoS ONE, 7: art.no. e48847, 2012.
* A numerical method for measuring symmetry in brain functional imaging data.
* https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3493608/
* https://dx.doi.org/10.1371/journal.pone.0048847
----------------------------------------------------------------------------
ZS Saad, SJ Gotts, K Murphy, G Chen, HJ Jo, A Martin, RW Cox.
Trouble at Rest: How Correlation Patterns and Group Differences Become
Distorted After Global Signal Regression. Brain Connectivity, 2012: 25-32.
* Another paper in the battle against Global Signal Regression.
* https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3484684/
* https://dx.doi.org/10.1089/brain.2012.0080
----------------------------------------------------------------------------
G Chen, ZS Saad, JC Britton, DS Pine, RW Cox
Linear mixed-effects modeling approach to FMRI group analysis.
NeuroImage, 73: 176-190, 2013.
* The math behind 3dLME.
* https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3404516/
* https://dx.doi.org/10.1016/j.neuroimage.2011.12.060
----------------------------------------------------------------------------
SJ Gotts, ZS Saad, HJ Jo, GL Wallace, RW Cox, A Martin.
The perils of global signal regression for group comparisons: A case study
of Autism Spectrum Disorders.
Frontiers in Human Neuroscience: art.no. 356, 2013.
* The long twilight struggle against Global Signal Regression continues.
* https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3709423/
* https://dx.doi.org/10.3389/fnhum.2013.00356
----------------------------------------------------------------------------
HJ Jo, SJ Gotts, RC Reynolds, PA Bandettini, A Martin, RW Cox, ZS Saad.
Effective preprocessing procedures virtually eliminate distance-dependent
motion artifacts in resting state FMRI.
Journal of Applied Mathematics: art.no. 935154, 2013.
* A reply to the Power 2012 paper on pre-processing resting state FMRI data,
showing how they got it wrong.
* https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3886863/
* https://dx.doi.org/10.1155/2013/935154
----------------------------------------------------------------------------
SJ Gotts, HJ Jo, GL Wallace, ZS Saad, RW Cox, A Martin.
Two distinct forms of functional lateralization in the human brain.
PNAS, 110: E3435-E3444, 2013.
* More about methodology and results for symmetry in brain function.
* https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3767540/
* https://dx.doi.org/10.1073/pnas.1302581110
----------------------------------------------------------------------------
ZS Saad, RC Reynolds, HJ Jo, SJ Gotts, G Chen, A Martin, RW Cox.
Correcting Brain-Wide Correlation Differences in Resting-State FMRI.
Brain Connectivity, 2013: 339-352.
* Just when you thought it was safe to go back into the waters of resting
state FMRI, another paper explaining why global signal regression is a
bad idea and a tentative step towards a different solution.
* https://www.ncbi.nlm.nih.gov/pubmed/23705677
* https://dx.doi.org/10.1089/brain.2013.0156
----------------------------------------------------------------------------
P Kundu, ND Brenowitz, V Voon, Y Worbe, PE Vertes, SJ Inati, ZS Saad,
PA Bandettini, ET Bullmore.
Integrated strategy for improving functional connectivity mapping using
multiecho fMRI. PNAS 110: 16187-16192, 2013.
* A data acquisition and processing strategy for improving resting state FMRI.
* https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3791700/
* https://dx.doi.org/10.1073/pnas.1301725110
----------------------------------------------------------------------------
PA Taylor, ZS Saad.
FATCAT: (An Efficient) Functional And Tractographic Connectivity Analysis
Toolbox. Brain Connectivity 3:523-535, 2013.
* Introducing diffusion-based tractography tools in AFNI, with particular
emphases on complementing FMRI analysis and in performing interactive
visualization with SUMA.
* https://www.ncbi.nlm.nih.gov/pubmed/23980912
* https://dx.doi.org/10.1089/brain.2013.0154
----------------------------------------------------------------------------
G Chen, NE Adleman, ZS Saad, E Leibenluft, RW Cox.
Applications of multivariate modeling to neuroimaging group analysis:
A comprehensive alternative to univariate general linear model.
NeuroImage 99:571-588, 2014.
* The fun stuff behind 3dMVM == more complex linear modeling for groups.
* https://dx.doi.org/10.1016/j.neuroimage.2014.06.027
* https://sscc.nimh.nih.gov/pub/dist/doc/papers/3dMVM_2014.pdf
----------------------------------------------------------------------------
Taylor PA, Chen G, Cox RW, Saad ZS.
Open Environment for Multimodal Interactive Connectivity
Visualization and Analysis. Brain Connectivity 6(2):109-21, 2016.
* Visualization and MVM stats tools using tracking (or even functional
connectivity).
* https://dx.doi.org/10.1089/brain.2015.0363
* https://sscc.nimh.nih.gov/pub/dist/papers/ASF_2015_draft_BCinpress.pdf
----------------------------------------------------------------------------
G Chen, Y-W Shin, PA Taylor, DR GLen, RC Reynolds, RB Israel, RW Cox.
Untangling the relatedness among correlations, part I: Nonparametric
approaches to inter-subject correlation analysis at the group level.
NeuroImage 142:248-259, 2016.
Proper statistical analysis (FPR control) when correlating FMRI time
series data amongst multiple subjects, using nonparametric methods.
* https://doi.org/10.1016/j.neuroimage.2016.05.023
----------------------------------------------------------------------------
G Chen, PA Taylor, Y-W Shin, RC Reynolds, RW Cox.
Untangling the relatedness among correlations, Part II: Inter-subject
correlation group analysis through linear mixed-effects modeling.
NeuroImage 147:825-840 2017.
* Just when you thought it was safe to go back into the brain data:
this time, using parametric methods.
* https://doi.org/10.1016/j.neuroimage.2016.08.029
----------------------------------------------------------------------------
G Chen, PA Taylor, X Qu, PJ Molfese, PA Bandettini, RW Cox, ES Finn.
Untangling the relatedness among correlations, part III: Inter-subject
correlation analysis through Bayesian multilevel modeling for naturalistic
scanning.
NeuroImage, 2019.
* https://doi.org/10.1016/j.neuroimage.2019.116474
* https://www.ncbi.nlm.nih.gov/pubmed/31884057
* https://www.biorxiv.org/content/10.1101/655738v1.full
----------------------------------------------------------------------------
RW Cox, G Chen, DR Glen, RC Reynolds, PA Taylor.
fMRI clustering and false-positive rates.
PNAS 114:E3370-E3371, 2017.
* Response to Eklund's (et al.) paper about clustering in PNAS 2016.
* https://arxiv.org/abs/1702.04846
* https://doi.org/10.1073/pnas.1614961114
----------------------------------------------------------------------------
RW Cox, G Chen, DR Glen, RC Reynolds, PA Taylor.
FMRI Clustering in AFNI: False Positive Rates Redux.
Brain Connectivity 7:152-171, 2017.
* A discussion of the cluster-size thresholding updates made to
AFNI in early 2017.
* https://arxiv.org/abs/1702.04845
* https://doi.org/10.1089/brain.2016.0475
----------------------------------------------------------------------------
S Song, RPH Bokkers, MA Edwardson, T Brown, S Shah, RW Cox, ZS Saad,
RC Reynolds, DR Glen, LG Cohen LG, LL Latour.
Temporal similarity perfusion mapping: A standardized and model-free method
for detecting perfusion deficits in stroke.
PLoS ONE 12, Article number e0185552, 2017.
* Applying AFNI's InstaCorr module to stroke perfusion mapping.
* https://doi.org/10.1371/journal.pone.0185552
* https://www.ncbi.nlm.nih.gov/pubmed/28973000
----------------------------------------------------------------------------
G Chen, PA Taylor, SP Haller, K Kircanski, J Stoddard, DS Pine, E Leibenluft,
Brotman MA, RW Cox.
Intraclass correlation: Improved modeling approaches and applications for
neuroimaging.
Human Brain Mapping, 39:1187-1206 2018.
* Discussion of ICC methods, and distinctions among them.
* https://doi.org/10.1002/hbm.23909
* https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5807222/
----------------------------------------------------------------------------
PA Taylor, G Chen, DR Glen, JK Rajendra, RC Reynolds, RW Cox.
FMRI processing with AFNI: Some comments and corrections on 'Exploring the
Impact of Analysis Software on Task fMRI Results'.
* https://www.biorxiv.org/content/10.1101/308643v1.abstract
* https://doi.org/10.1101/308643
----------------------------------------------------------------------------
RW Cox.
Equitable Thresholding and Clustering: A Novel Method for Functional
Magnetic Resonance Imaging Clustering in AFNI.
Brain Connectivity 9:529-538, 2019.
* https://doi.org/10.1089/brain.2019.0666
----------------------------------------------------------------------------
G Chen, RW Cox, DR Glen, JK Rajendra, RC Reynolds, PA Taylor.
A tail of two sides: Artificially doubled false positive rates in
neuroimaging due to the sidedness choice with t-tests.
Human Brain Mapping 40:1037-1043, 2019.
* https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6328330/
* https://dx.doi.org/10.1002/hbm.24399
----------------------------------------------------------------------------
G Chen, Y Xiao, PA Taylor, JK Rajendra, T Riggins, F Geng, E Redcay, RW Cox.
Handling Multiplicity in Neuroimaging Through Bayesian Lenses with
Multilevel Modeling.
Neuroinformatics 17:515-545, 2019.
* https://link.springer.com/article/10.1007/s12021-018-9409-6
* https://www.biorxiv.org/content/10.1101/238998v1.abstract
----------------------------------------------------------------------------
DR Glen, PA Taylor, BR Buchsbaum, RW Cox, and RC Reynolds.
Beware (Surprisingly Common) Left-Right Flips in Your MRI Data:
An Efficient and Robust Method to Check MRI Dataset Consistency Using AFNI.
Frontiers in Neuroinformatics, 25 May 2020.
* https://doi.org/10.3389/fninf.2020.00018
* https://www.medrxiv.org/content/10.1101/19009787v4
----------------------------------------------------------------------------
V Roopchansingh, JJ French Jr, DM Nielson, RC Reynolds, DR Glen, P D’Souza,
PA Taylor, RW Cox, AE Thurm.
EPI Distortion Correction is Easy and Useful, and You Should Use It:
A case study with toddler data.
* https://www.biorxiv.org/content/10.1101/2020.09.28.306787v1
----------------------------------------------------------------------------
G Chen, TA Nash, KM Cole, PD Kohn, S-M Wei, MD Gregory, DP Eisenberg,
RW Cox, KF Berman, JS Kippenham.
Beyond linearity in neuroimaging: Capturing nonlinear relationships with
application to longitudinal studies.
* https://doi.org/10.1016/j.neuroimage.2021.117891
* https://pubmed.ncbi.nlm.nih.gov/33667672/
----------------------------------------------------------------------------
POSTERS on varied subjects from the AFNI development group can be found at
* https://afni.nimh.nih.gov/sscc/posters
------------------------------------------------------------------------------------
SLIDE IMAGES to help with learning the AFNI GUI
https://afni.nimh.nih.gov/pub/dist/doc/program_help/images/afni03/
------------------------------------------------------------------------------------
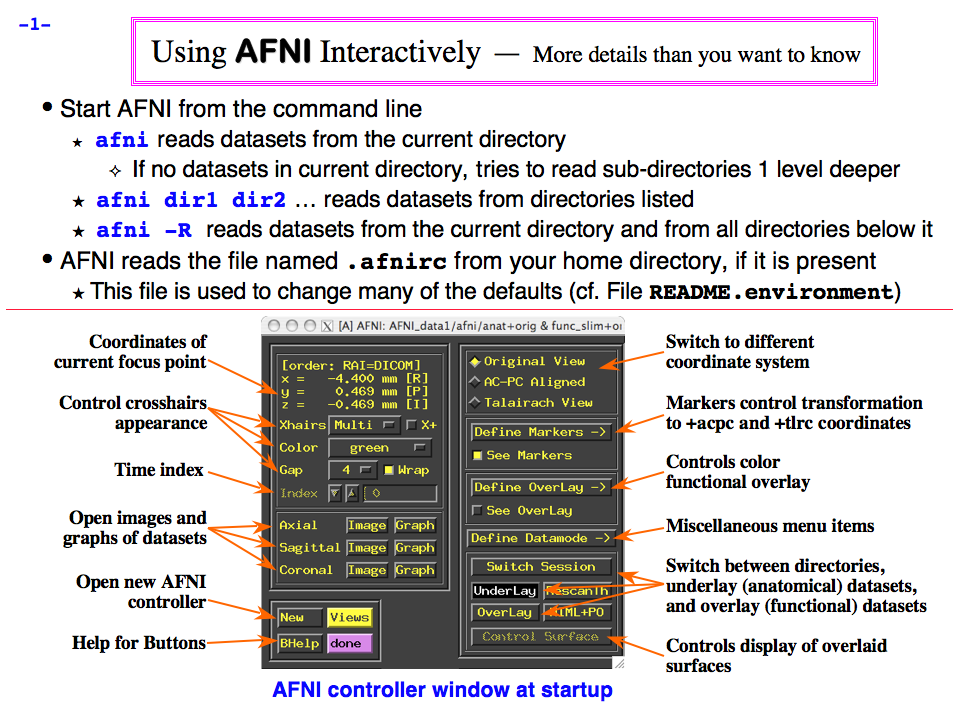
https://afni.nimh.nih.gov/pub/dist/doc/program_help/images/afni03/Slide01.png
------------------------------------------------------------------------------------

https://afni.nimh.nih.gov/pub/dist/doc/program_help/images/afni03/Slide02.png
------------------------------------------------------------------------------------

https://afni.nimh.nih.gov/pub/dist/doc/program_help/images/afni03/Slide03.png
------------------------------------------------------------------------------------
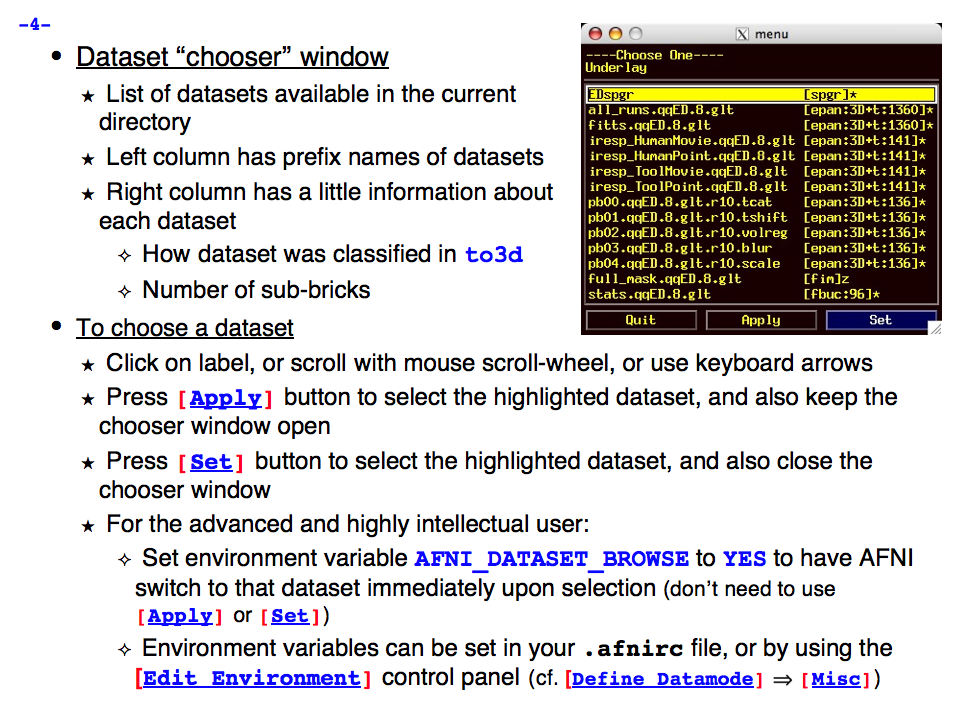
https://afni.nimh.nih.gov/pub/dist/doc/program_help/images/afni03/Slide04.png
------------------------------------------------------------------------------------

https://afni.nimh.nih.gov/pub/dist/doc/program_help/images/afni03/Slide05.png
------------------------------------------------------------------------------------

https://afni.nimh.nih.gov/pub/dist/doc/program_help/images/afni03/Slide06.png
------------------------------------------------------------------------------------
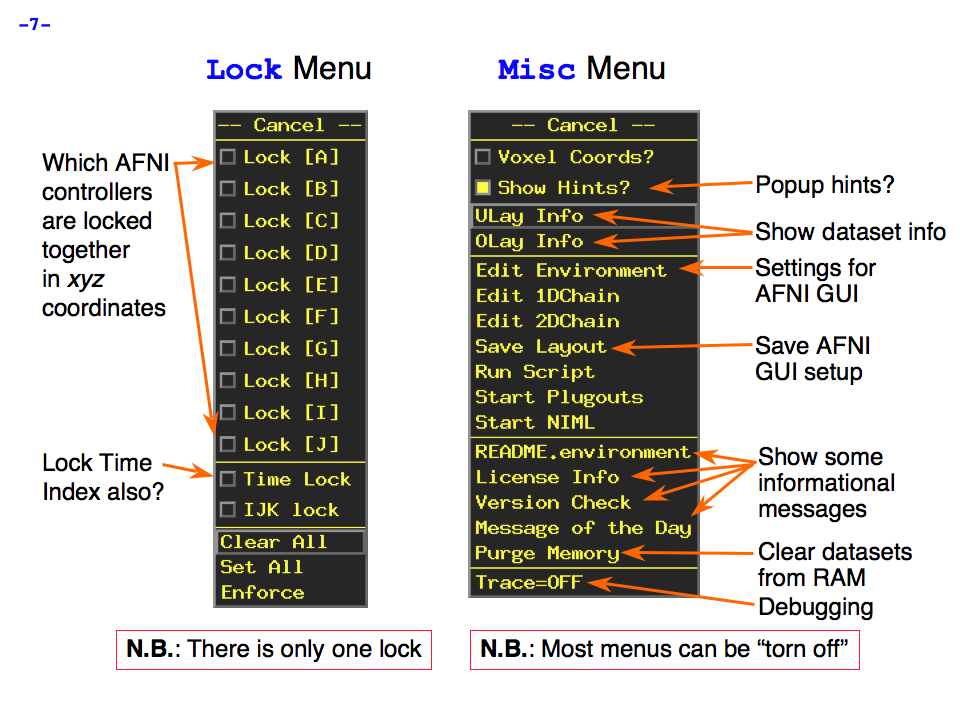
https://afni.nimh.nih.gov/pub/dist/doc/program_help/images/afni03/Slide07.png
------------------------------------------------------------------------------------
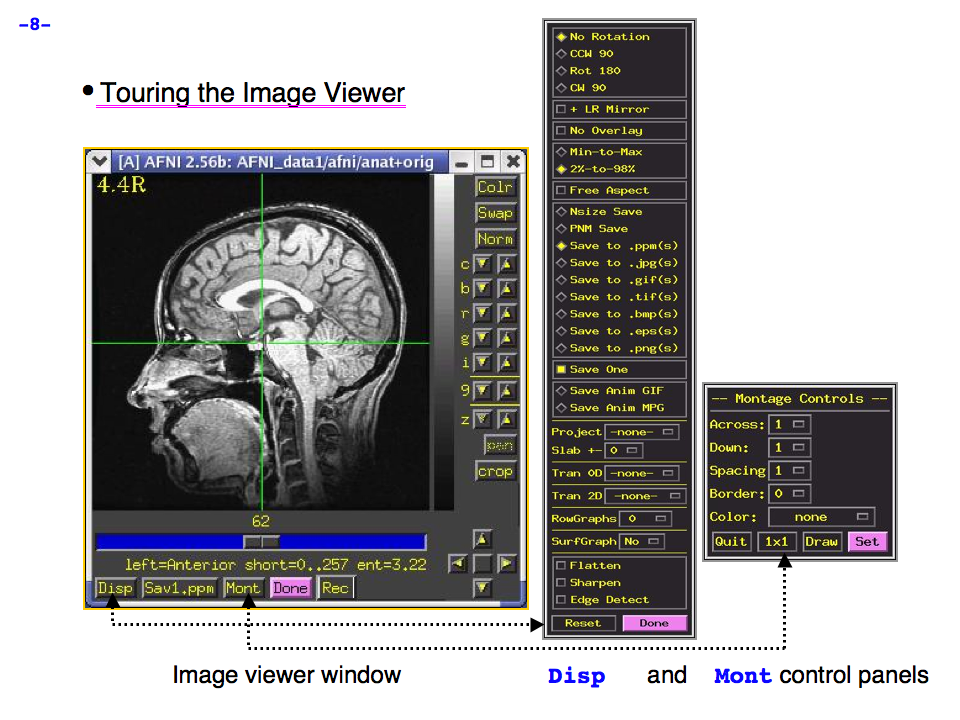
https://afni.nimh.nih.gov/pub/dist/doc/program_help/images/afni03/Slide08.png
------------------------------------------------------------------------------------

https://afni.nimh.nih.gov/pub/dist/doc/program_help/images/afni03/Slide09.png
------------------------------------------------------------------------------------

https://afni.nimh.nih.gov/pub/dist/doc/program_help/images/afni03/Slide10.png
------------------------------------------------------------------------------------
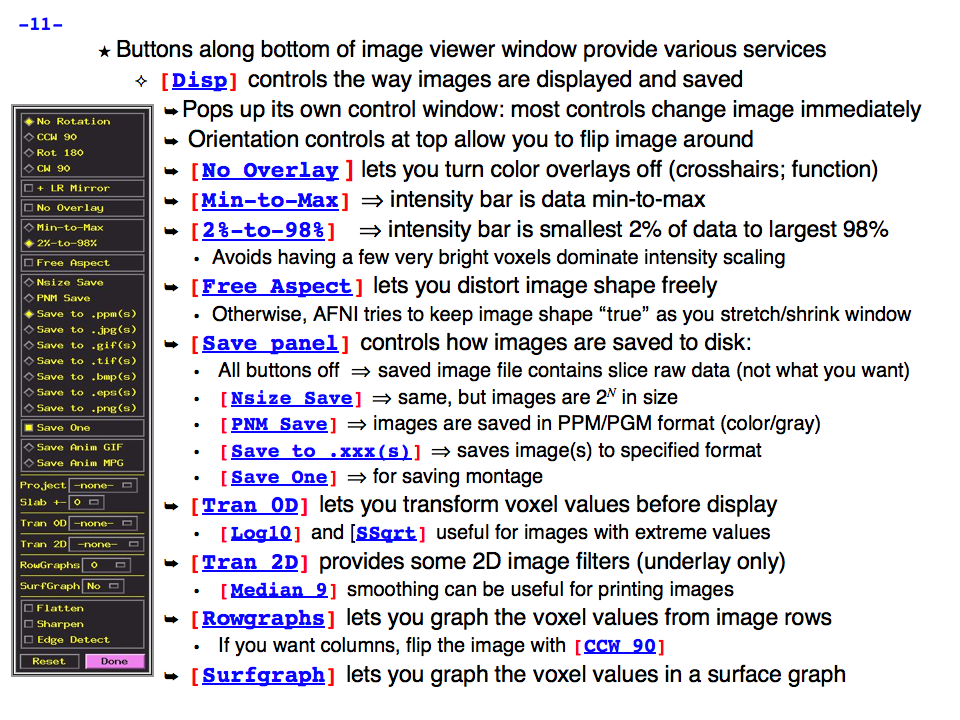
https://afni.nimh.nih.gov/pub/dist/doc/program_help/images/afni03/Slide11.png
------------------------------------------------------------------------------------

https://afni.nimh.nih.gov/pub/dist/doc/program_help/images/afni03/Slide12.png
------------------------------------------------------------------------------------

https://afni.nimh.nih.gov/pub/dist/doc/program_help/images/afni03/Slide13.png
------------------------------------------------------------------------------------

https://afni.nimh.nih.gov/pub/dist/doc/program_help/images/afni03/Slide14.png
------------------------------------------------------------------------------------

https://afni.nimh.nih.gov/pub/dist/doc/program_help/images/afni03/Slide15.png
------------------------------------------------------------------------------------

https://afni.nimh.nih.gov/pub/dist/doc/program_help/images/afni03/Slide16.png
------------------------------------------------------------------------------------

https://afni.nimh.nih.gov/pub/dist/doc/program_help/images/afni03/Slide17.png
------------------------------------------------------------------------------------

https://afni.nimh.nih.gov/pub/dist/doc/program_help/images/afni03/Slide18.png
------------------------------------------------------------------------------------
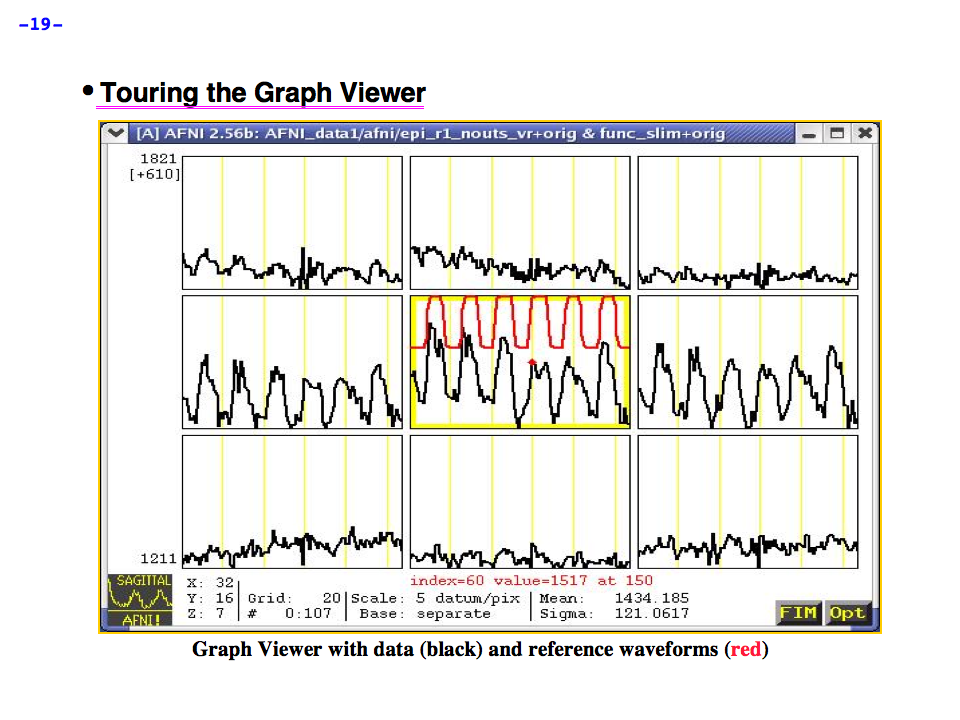
https://afni.nimh.nih.gov/pub/dist/doc/program_help/images/afni03/Slide19.png
------------------------------------------------------------------------------------
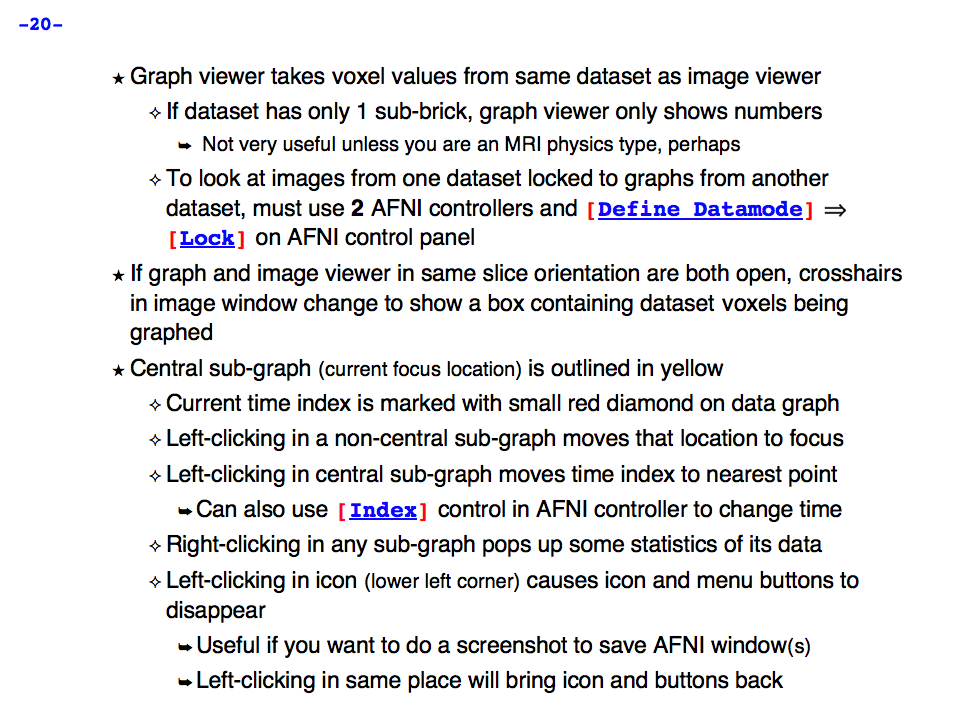
https://afni.nimh.nih.gov/pub/dist/doc/program_help/images/afni03/Slide20.png
------------------------------------------------------------------------------------

https://afni.nimh.nih.gov/pub/dist/doc/program_help/images/afni03/Slide21.png
------------------------------------------------------------------------------------

https://afni.nimh.nih.gov/pub/dist/doc/program_help/images/afni03/Slide22.png
------------------------------------------------------------------------------------

https://afni.nimh.nih.gov/pub/dist/doc/program_help/images/afni03/Slide23.png
------------------------------------------------------------------------------------
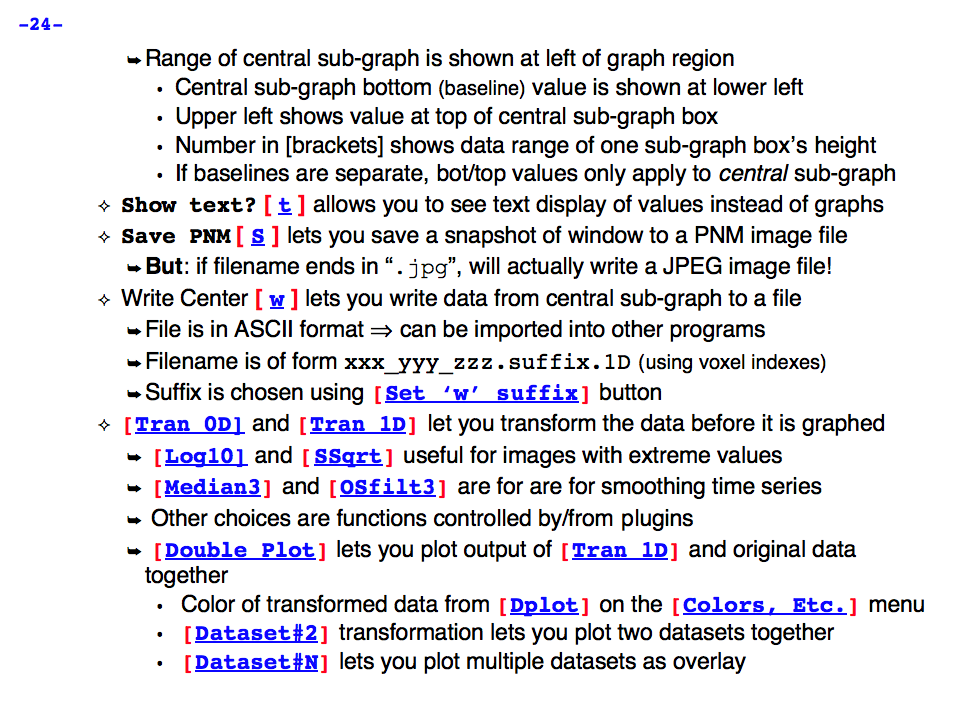
https://afni.nimh.nih.gov/pub/dist/doc/program_help/images/afni03/Slide24.png
------------------------------------------------------------------------------------
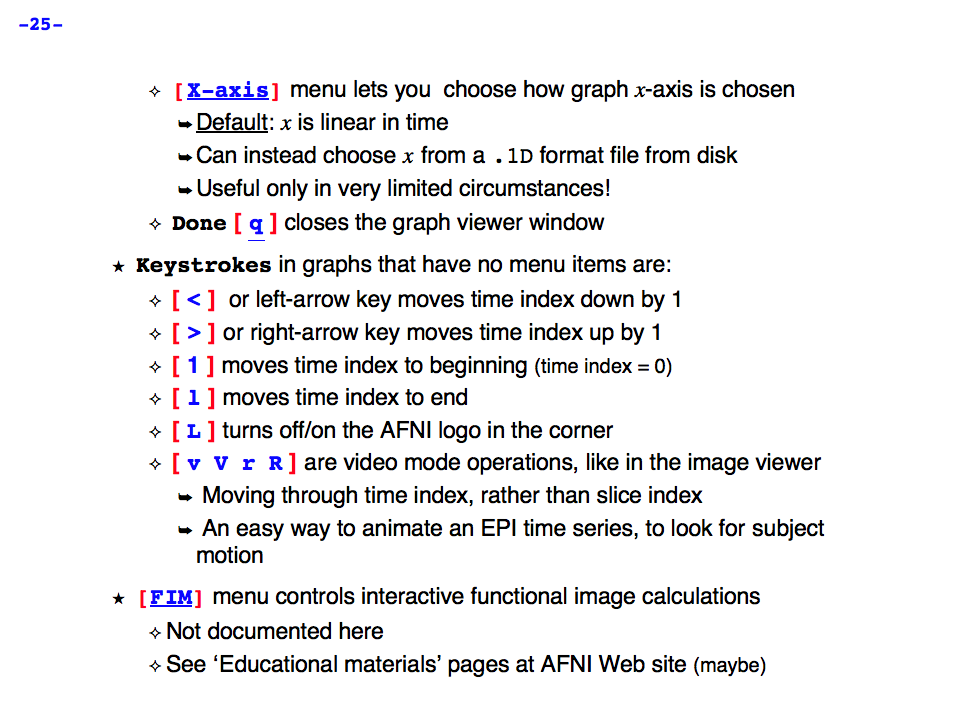
https://afni.nimh.nih.gov/pub/dist/doc/program_help/images/afni03/Slide25.png
------------------------------------------------------------------------------------
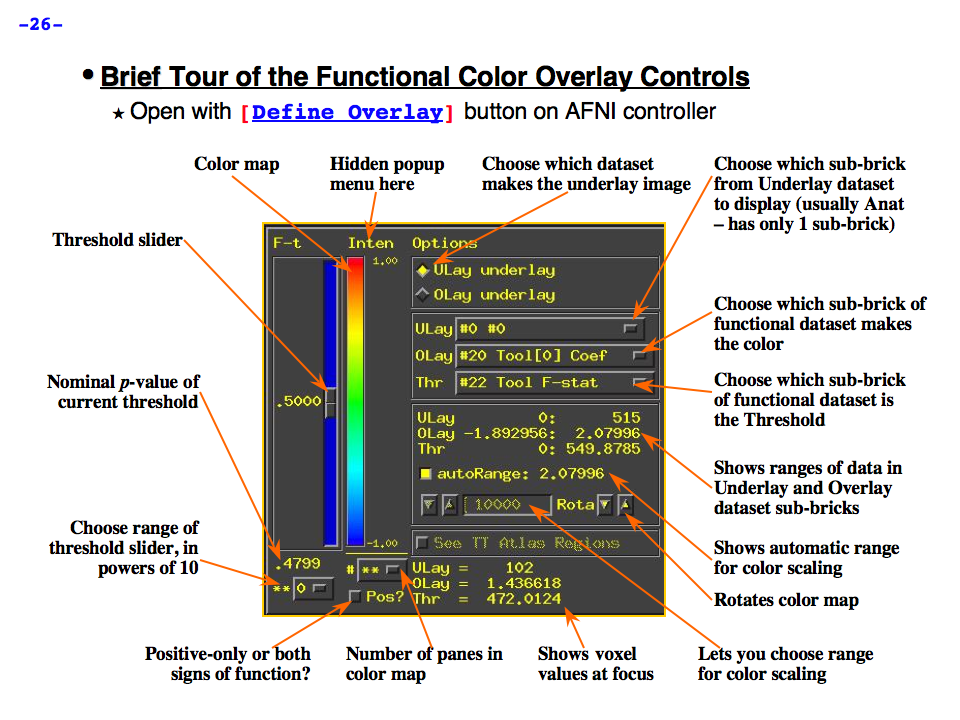
https://afni.nimh.nih.gov/pub/dist/doc/program_help/images/afni03/Slide26.png
------------------------------------------------------------------------------------

https://afni.nimh.nih.gov/pub/dist/doc/program_help/images/afni03/Slide27.png
------------------------------------------------------------------------------------
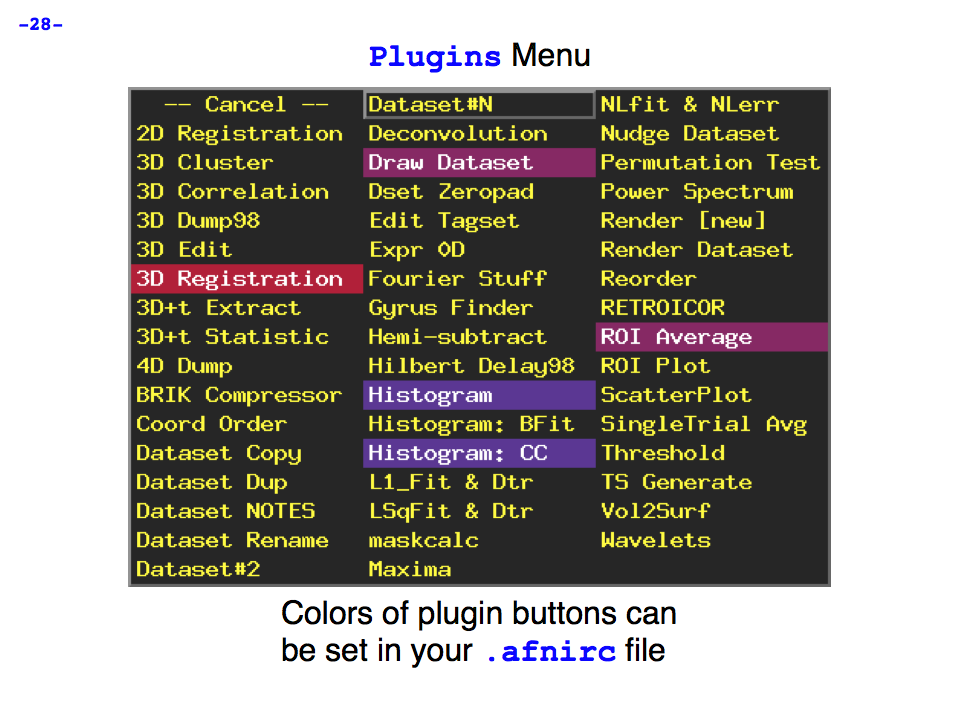
https://afni.nimh.nih.gov/pub/dist/doc/program_help/images/afni03/Slide28.png
------------------------------------------------------------------------------------
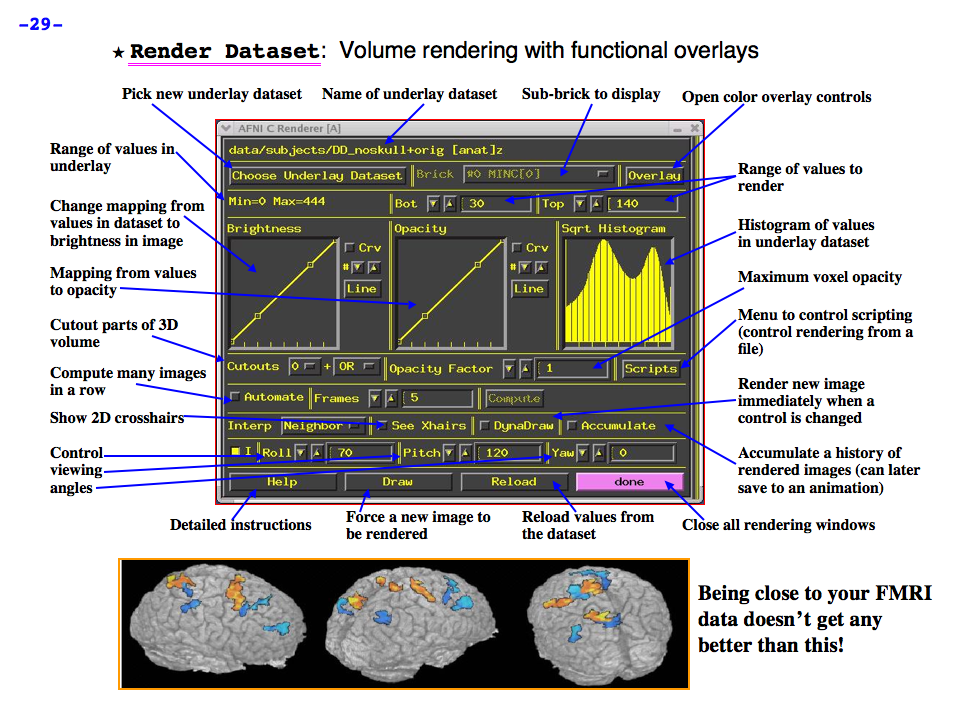
https://afni.nimh.nih.gov/pub/dist/doc/program_help/images/afni03/Slide29.png
------------------------------------------------------------------------------------

https://afni.nimh.nih.gov/pub/dist/doc/program_help/images/afni03/Slide30.png
------------------------------------------------------------------------------------

https://afni.nimh.nih.gov/pub/dist/doc/program_help/images/afni03/Slide31.png
------------------------------------------------------------------------------------

https://afni.nimh.nih.gov/pub/dist/doc/program_help/images/afni03/Slide32.png
------------------------------------------------------------------------------------
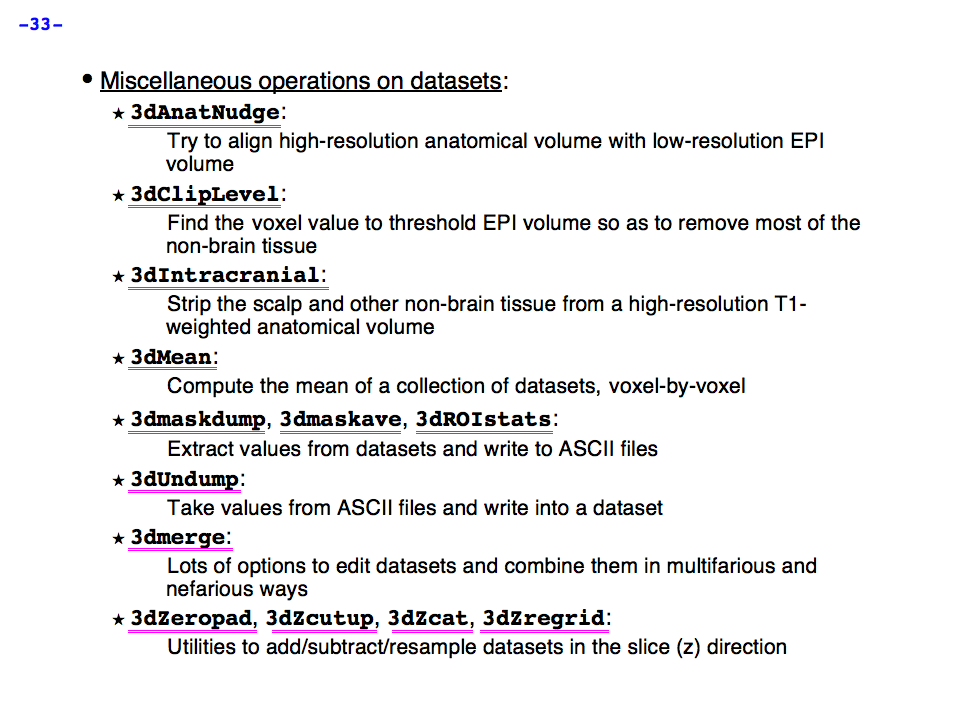
https://afni.nimh.nih.gov/pub/dist/doc/program_help/images/afni03/Slide33.png
------------------------------------------------------------------------------------

https://afni.nimh.nih.gov/pub/dist/doc/program_help/images/afni03/Slide34.png
------------------------------------------------------------------------------------
This page auto-generated on
Mon Feb 23 10:14:13 PM EST 2026


































