9.1.1. Task FMRI processing: the MACAQUE_DEMO¶
Overview¶
Welcome to the homepage of the demo for processing task FMRI macaque data.
The demo dataset contains an anatomical dataset and several EPIs collected during a visual stimulus with four stimulus classes related to specific faces and objects shown to the subject.
The processing scripts include calculating nonlinear alignment between
the subject’s anatomical (using AFNI’s @animal_warper) and the
stereotaxic NMT template. Additionally, a full single subject
processing pipeline is constructed with afni_proc.py, including an
HRF appropriate for this EPI data that had been collected with MION.
These scripts are meant to providing and example and starting point of task processing. Many features can (and, in general, will) be adjusted for your own study’s design. Please feel free to ask question on the AFNI Message Board, even with regards to setting up your study.
Thanks go to Adam Messinger, Ben Jung and Jakob Seidlitz for both the accompanying macaque data set and many processing suggestions/advice.
Download the MACAQUE_DEMO¶
To get the demo, copy+paste:
@Install_MACAQUE_DEMO
This will both download and unpack the demo in the directory where the command is run.
There is a README.txt describing the contents.
Demo contents¶
Input datasets¶
The demo contains one subject’s initial data:
- a standard, high-res anatomical (T1w) dataset
- a set of task FMRI (EPI) datasets; these are raw EPIs, with “place correction” already performed
- a set of stimulus timing files; these have already been adjusted for the TR removal that will be performed during processing
- reference template and atlas: here, the stereotaxic NMT and the D99 template in that space
Processing: nonlinear alignment with @animal_warper¶
Some processed datasets are also included. In particular, AFNI’s
@animal_warper script was run on the anatomical dataset to perform
both skullstripping and nonlinear alignment to template space (here,
the stereotaxic NMT atlas). This has been performed with the included
s00*.tcsh script. Some of the functionality and output of this
step can be seen in the automatically generated QC images:
| Alignment check: warped anatomical overlaying reference base edges |
|---|
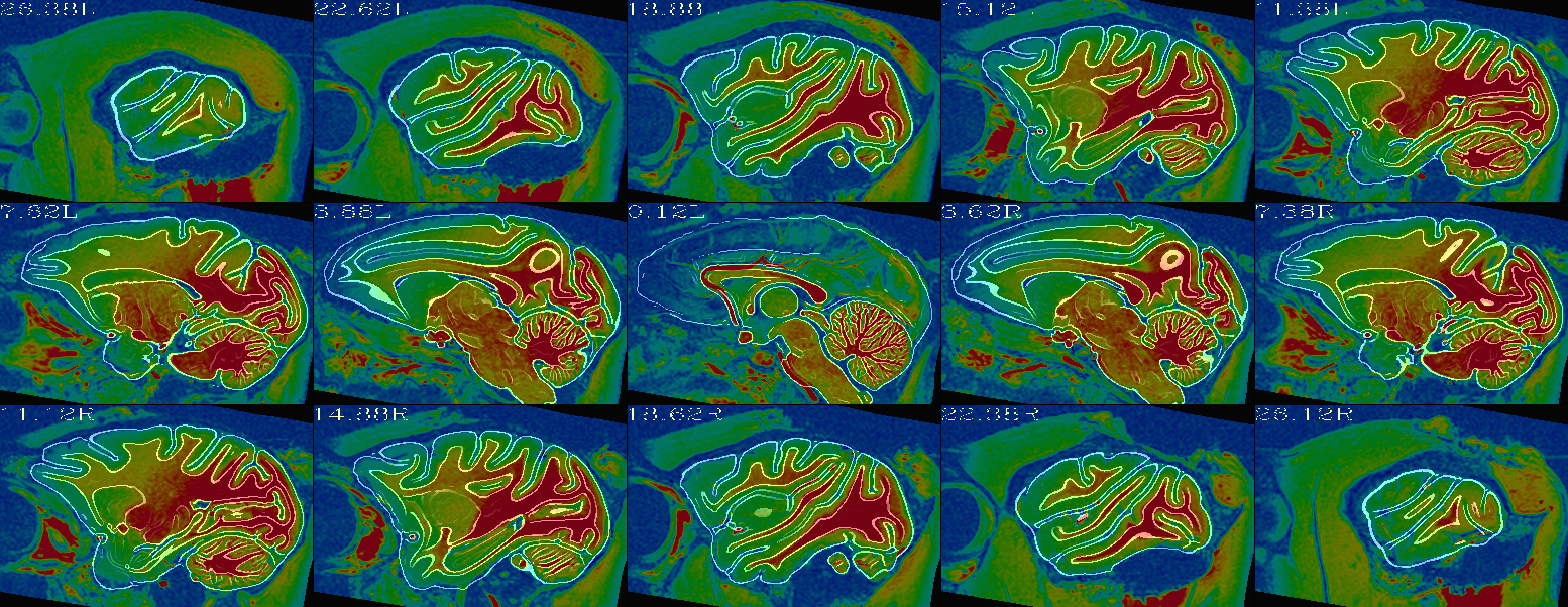
|
| Skullstripping check: native space anatomical underlaying brain mask |
|---|
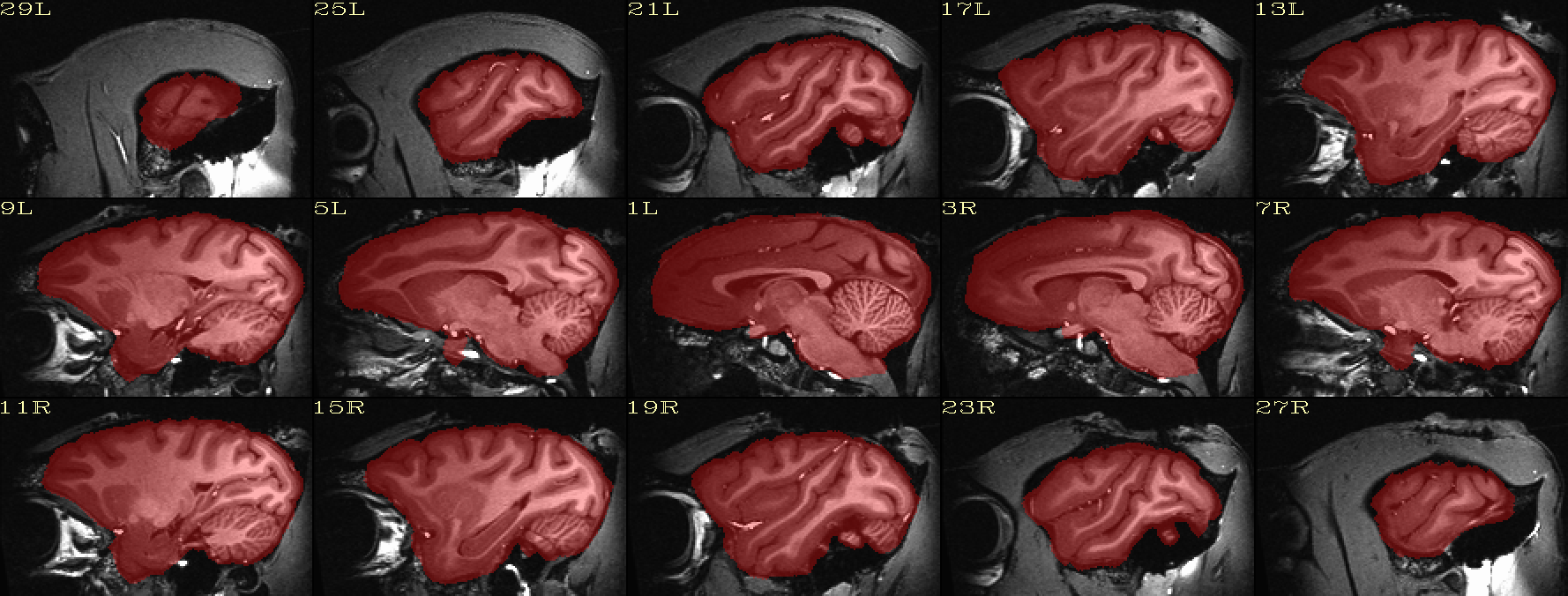
|
| Atlas ROIs mapped to native space |
|---|
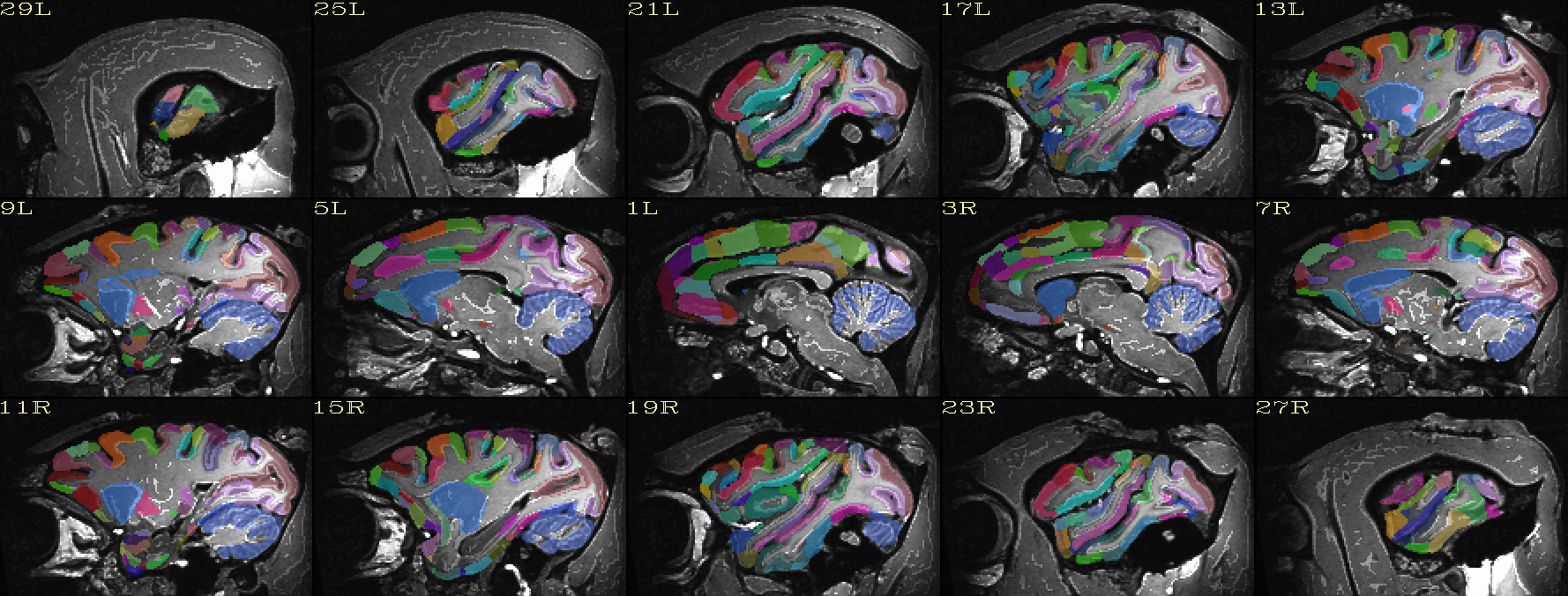
|
| SUMA view of ROI parcellation in native space (some ROIs have been made nearly transparent to show show inner regions) |
|---|
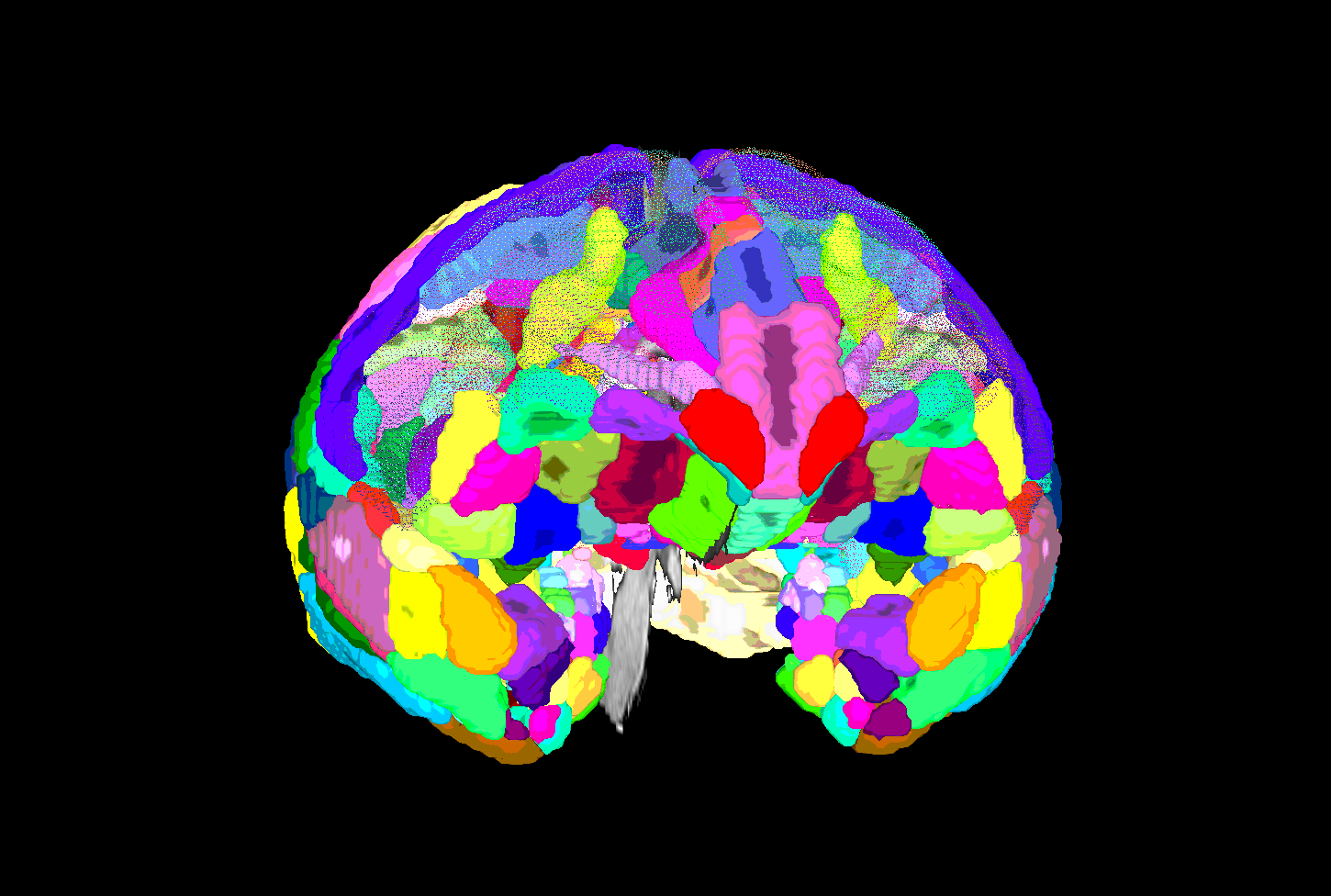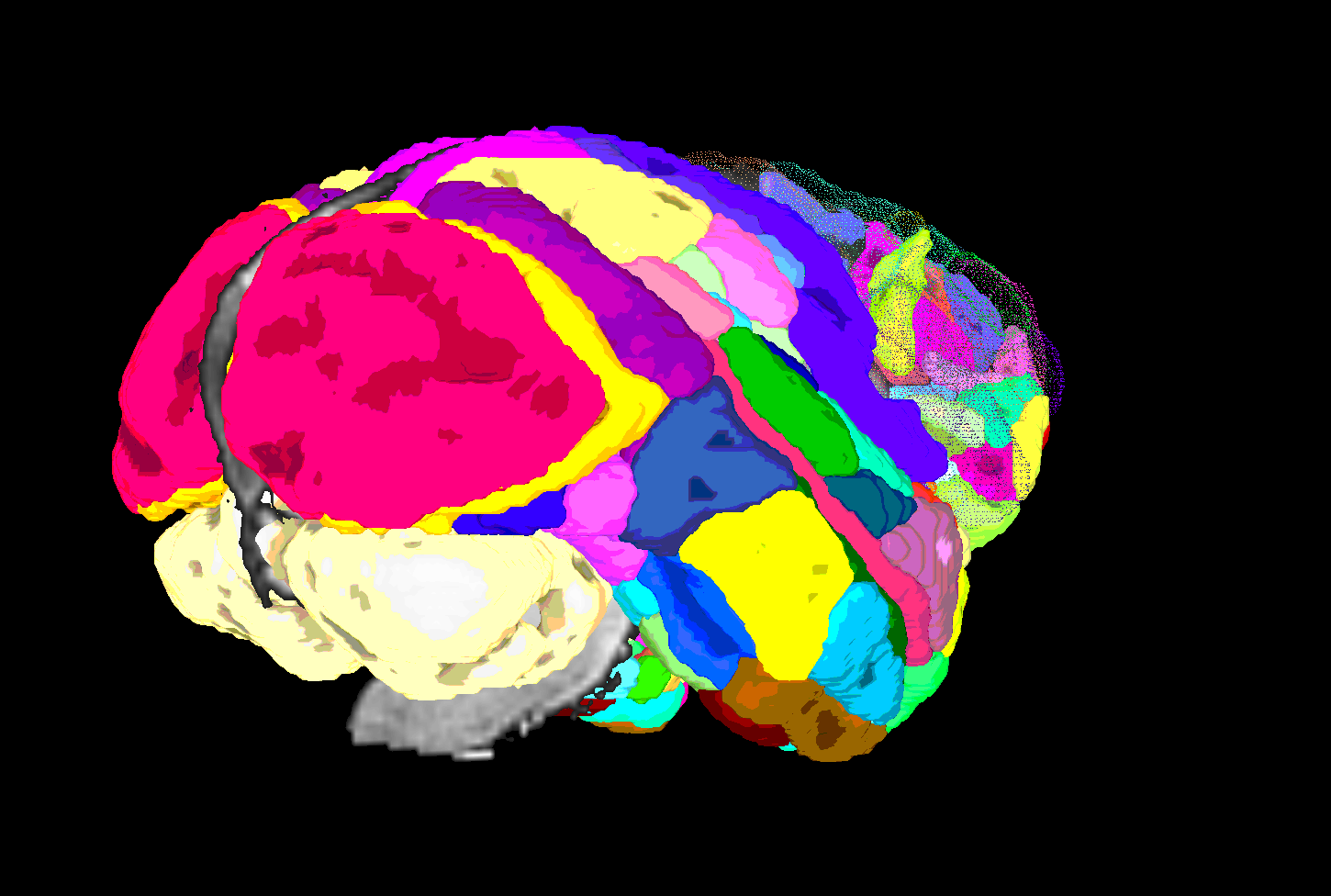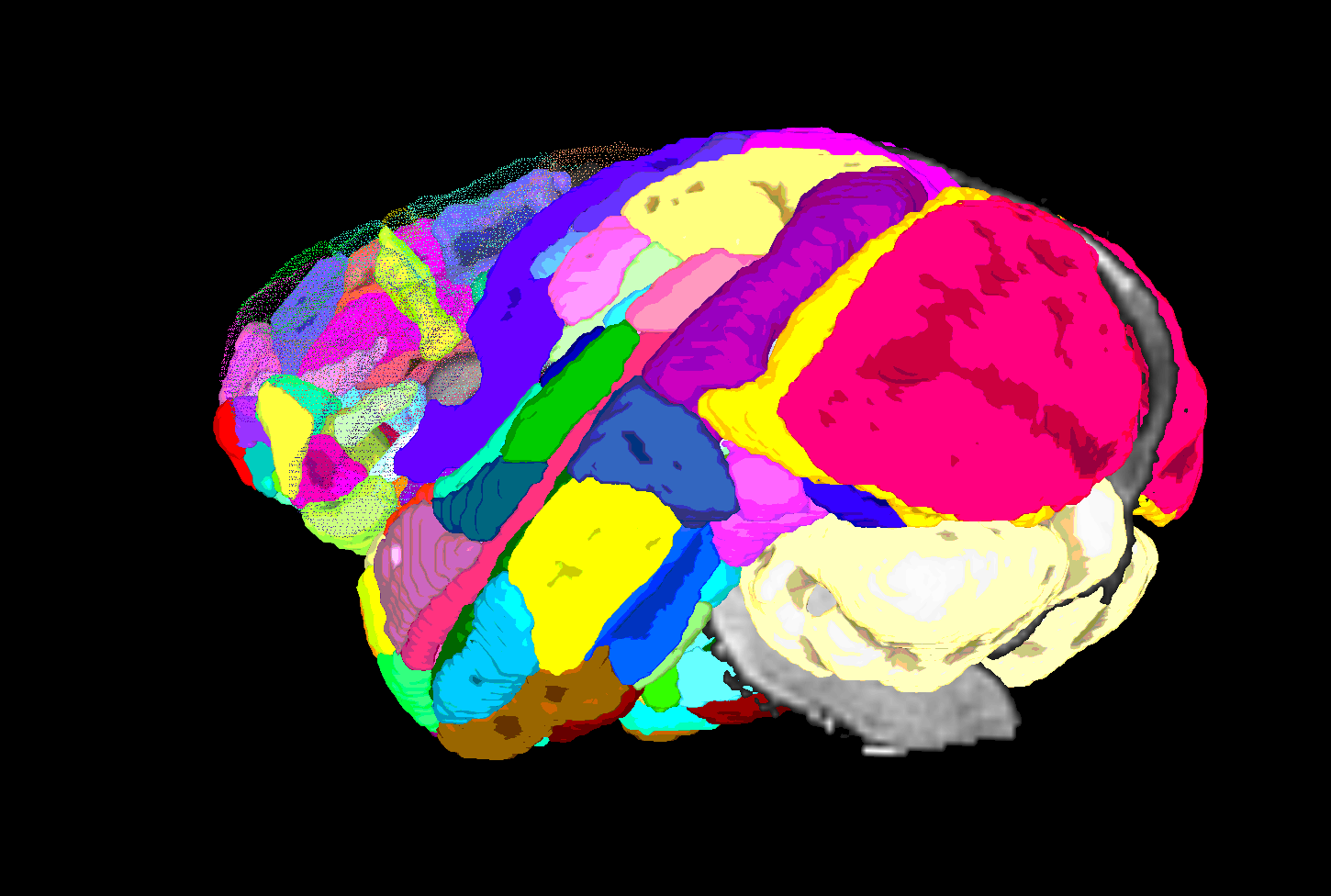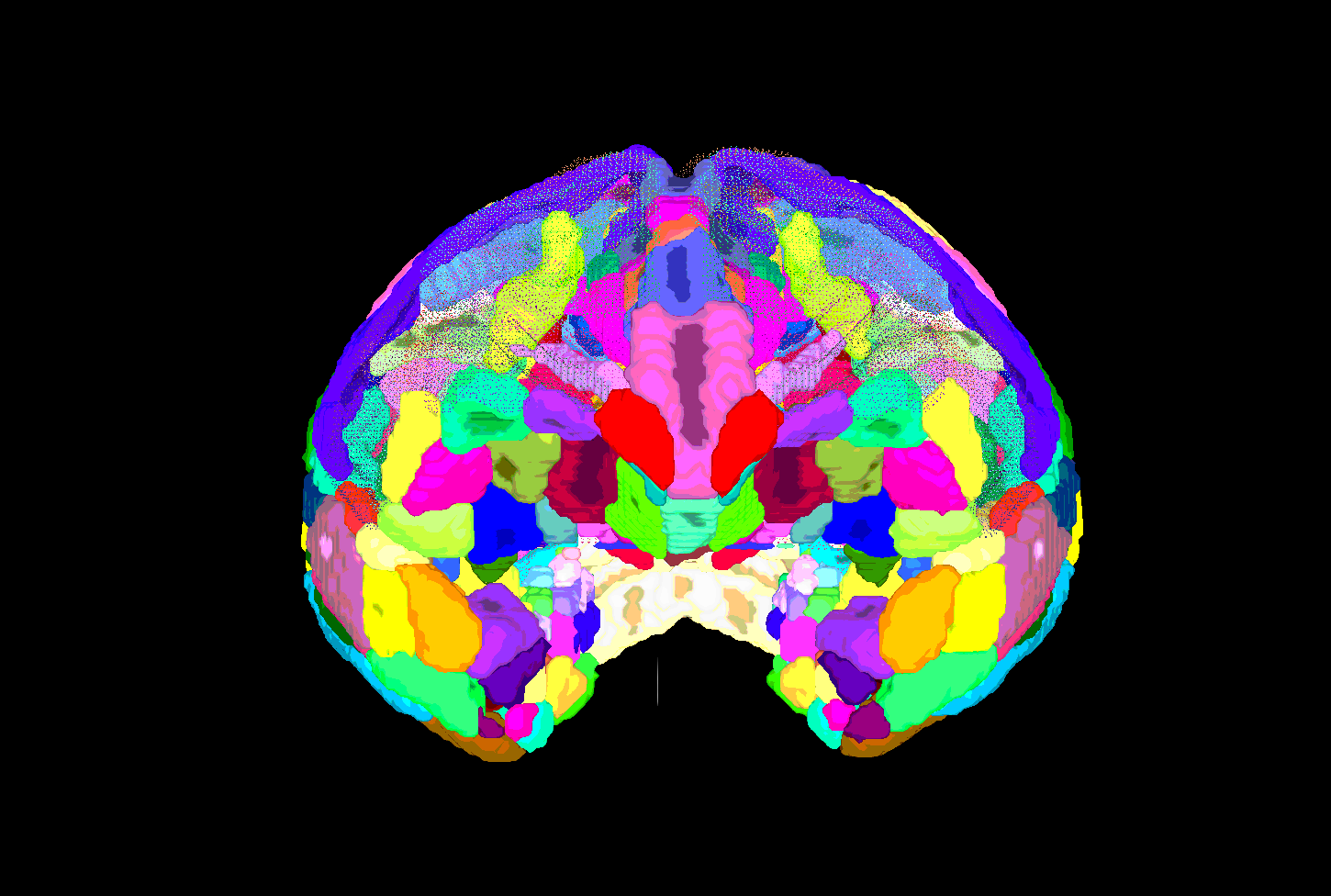
|
Outputs from this step are included as inputs for afni_proc.py
Processing: full single subject preprocessing with afni_proc.py¶
A full preprocessing script for the subject, including motion
correction and regression modeling, is generated using
afni_proc.py. The s01*.tcsh script contains an example of
including 4 EPI datasets for processing, and the s11*.tcsh script
runs over all 15 EPIs included in the demo (the latter takes notably
longer to run).
The outputs of the the afni_proc.py-generated scripts are not
included, but their automatically generated QC HTML files are.
| QC blocks: anat-to-template and stat modeling (full F-stat) | QC block: motion and outliers |
|---|---|
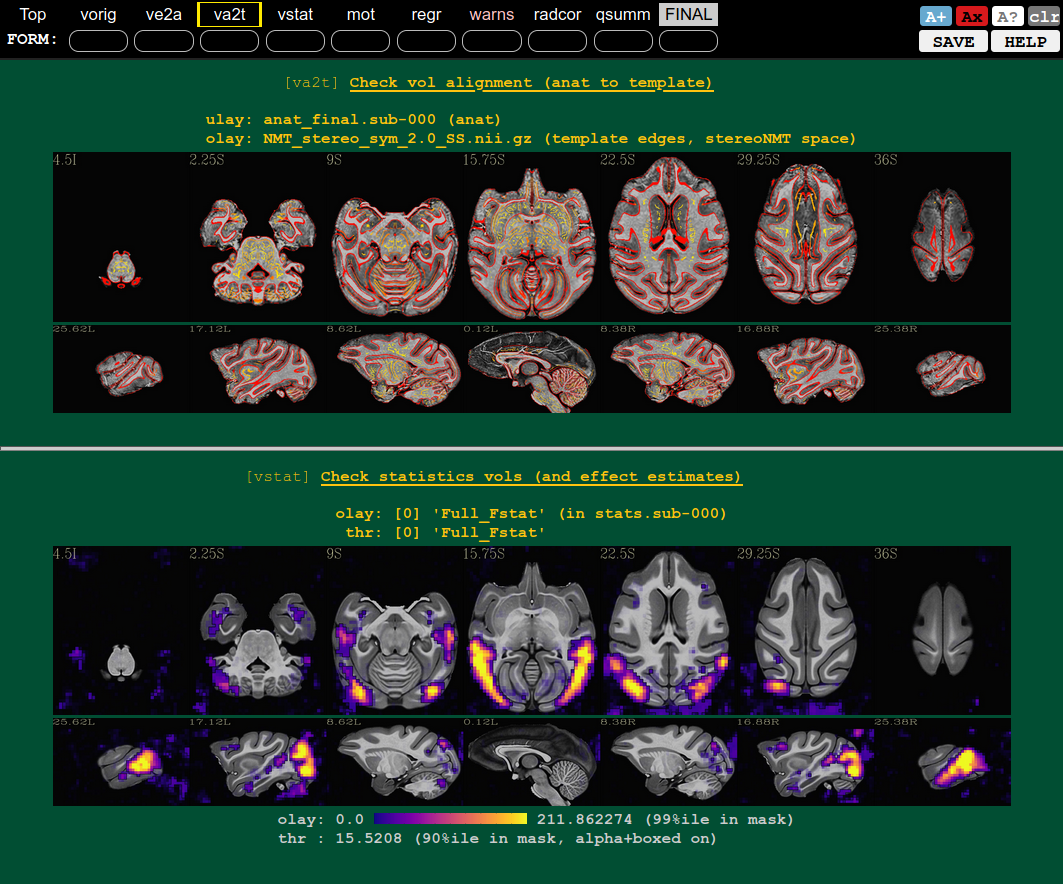
|
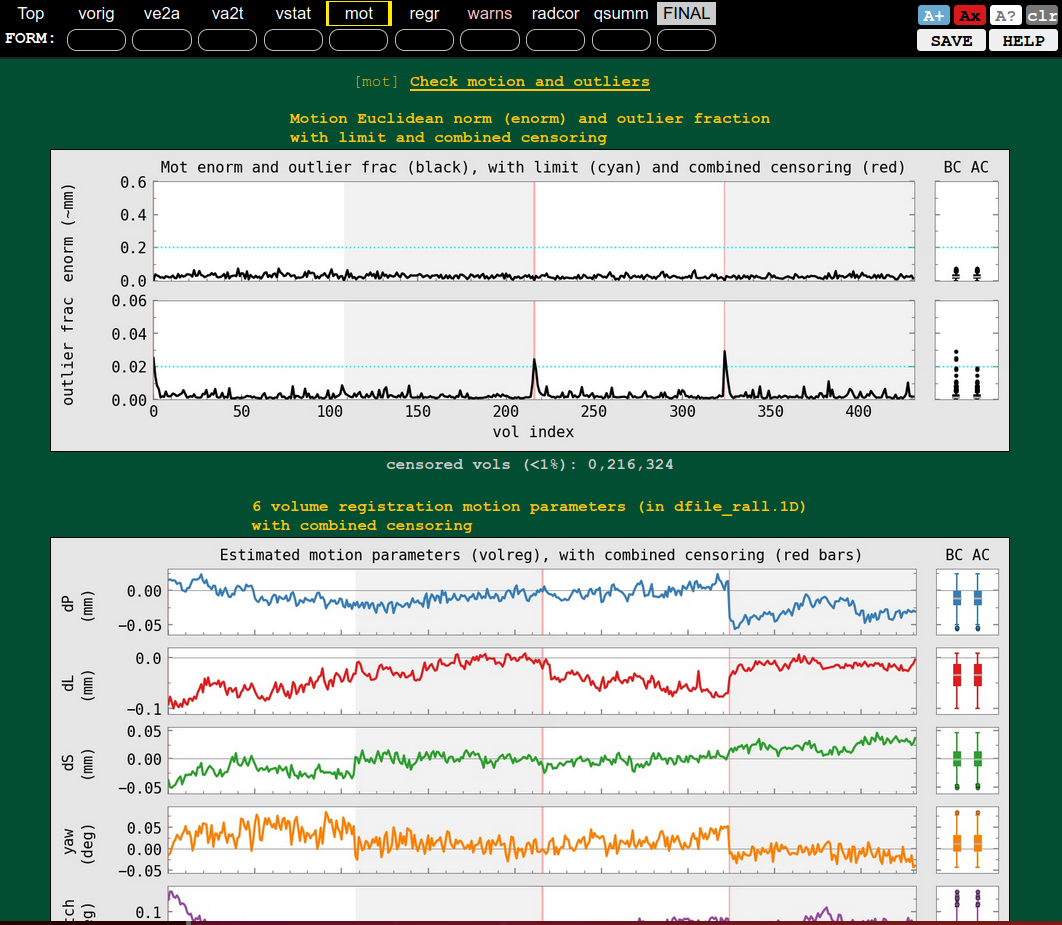
|
| QC block: warnings (collinearity, censoring, etc.) | QC block: regression (ideal response, DF counts, etc.) |
|---|---|
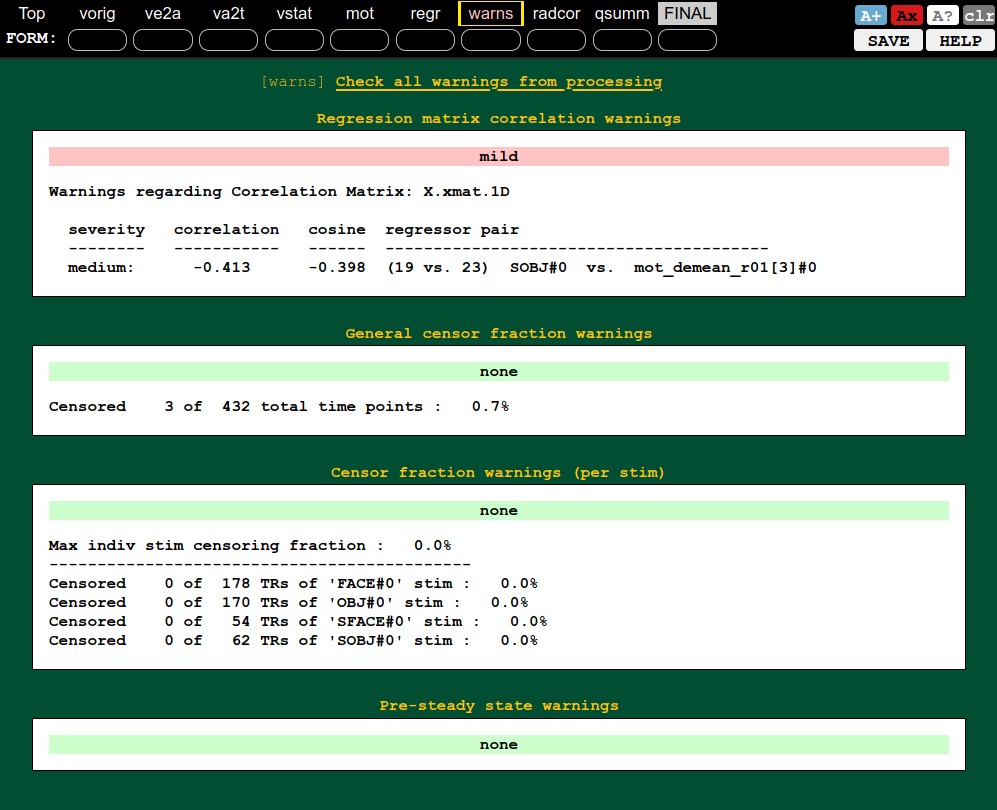
|
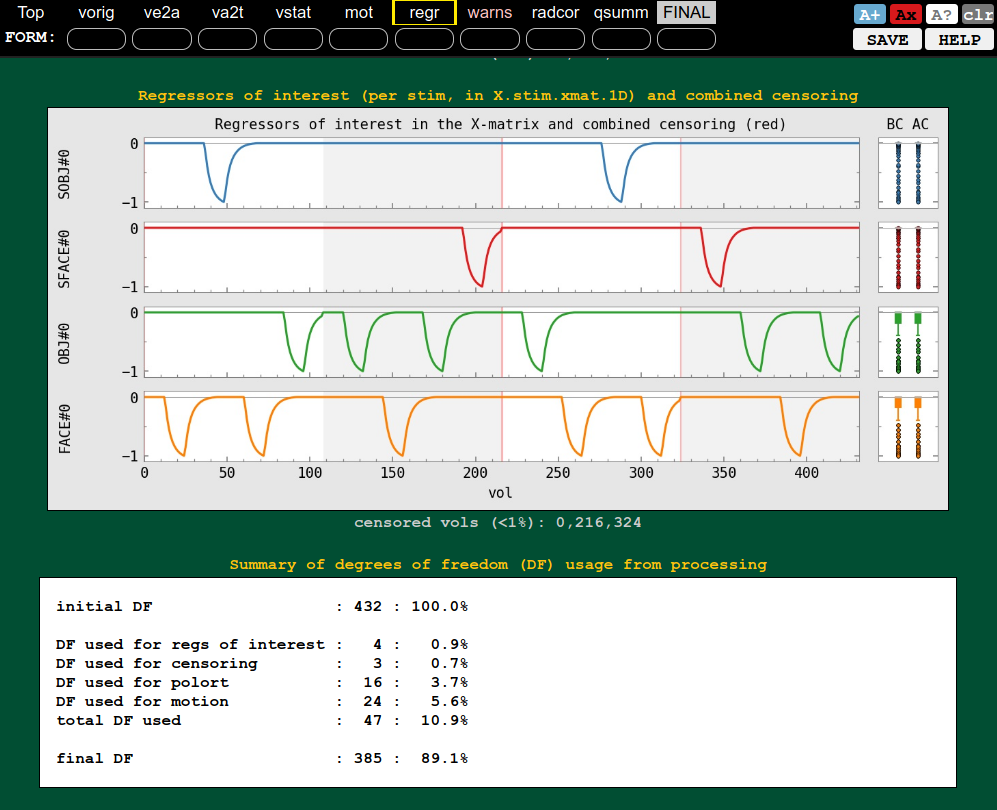
|
