11.1.3. Macaque template: NMT v1 (older)¶
Overview¶
Here we present the NIMH Macaque Template (NMT), version 1.2 and 1.3. This is a group template from 31 macaques with surfaces and GM/WM/CSF segmentations.
Please note that there is a newer version of this template. See this page: Macaque template: NMT v2 (current).
Note
NMT v1.* and NMT v2 are not compatible. They do not overlay, and they define different spaces. Please see Jung et al. (2020) for more details about their different properties.
NMT v1.3¶
Contents¶
NMT v1.3 contents |
Description |
|---|---|
CustomAtlases.niml |
atlas description text file for AFNI |
D99_atlas_1.2a_al2NMT.nii.gz |
D99 atlas in NMT v1.3 space |
NMT_brainmask.nii.gz |
binary brain mask (including cerebellum) |
NMT_env.csh |
script to set AFNI environment variables for AFNI to use
atlases and make AFNI more monkey friendly. Note: running
this script will change your default atlases for |
NMT_GM_cortical_mask.nii.gz |
binary mask of cortical GM |
NMT_GM_cortical_mask_withWM.nii.gz |
map of cortical GM and WM |
NMT.nii.gz |
the NMT itself, including non-brain material |
NMT_segmentation_4class.nii.gz |
map of tissue segmentation, cortex and cerebellum |
NMT_SS.nii.gz |
the NMT itself, skullstripped |
NMT_subject_align |
a script used to perform affine and nonlinear alignments of an
anatomical volume to the NMT using AFNI commands. Note: In
the NMT v2, this script has been replaced with the AFNI command
|
NMT_subject_morph |
a script that uses ANTs to perform template-based morphometrics of a native scan. Requires outputs from NMT_subject_align. Note: we do not recommend use of this script, as the new CIVET-macaque pipeline conducts automated surface-based morphometrics in the NMT space. |
NMT_subject_pipeline |
a wrapper script that performs NMT_subject_align, NMT_subject_process and NMT_subject_morph using a single command! |
NMT_subject_process |
a script used to perform template-based skullstripping and
segmentation in the native space of a subject using AFNI and
ANTs commands. Requires outputs from
NMT_subject_align. Note: In the NMT v2, template-based
skullstripping is performed by the AFNI command
|
processing_masks/ |
directory of tissue and segmentation volumes |
quick_view.csh |
example script to quickly view surfaces and volumes in |
README.md |
go ahead, I dare you to read it |
supplemental_D99/ |
directory of a table of D99 ROI index/label definitions |
supplemental_masks/ |
directory of NIFTI volumes of “other” structures (vasculature, cerebellum, LR brainmask, olfactory bulb) |
surfaces/ |
directory of GIFTI surfaces of many of the tissue/segmentation regions |
Example images¶
Full NMT with brainmask |
|---|
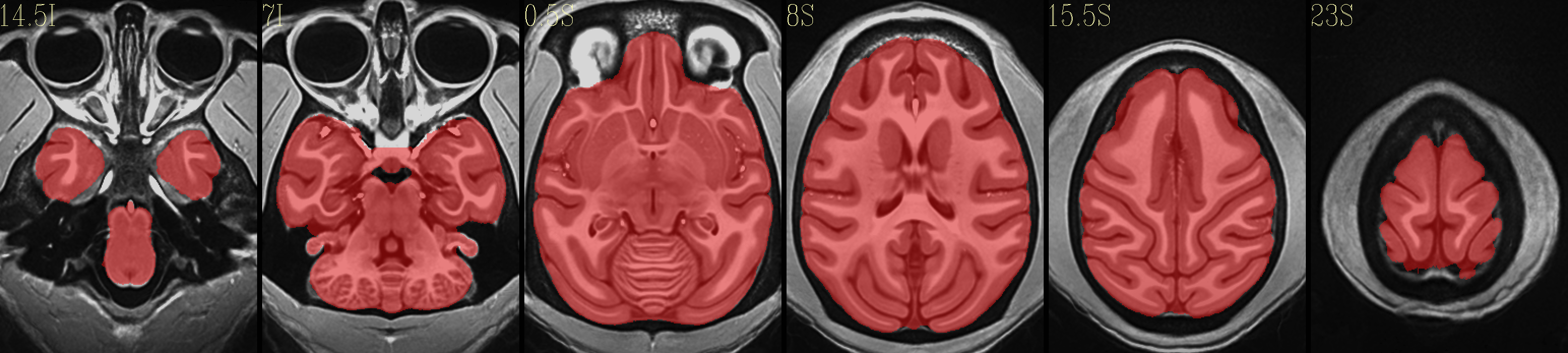
|
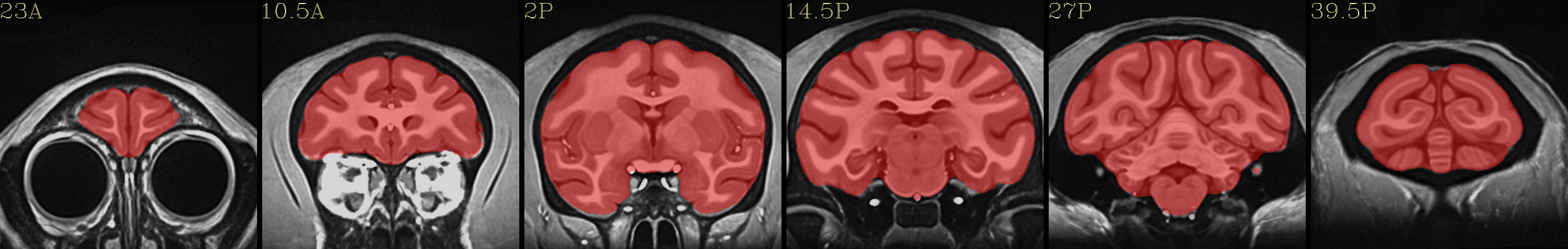
|
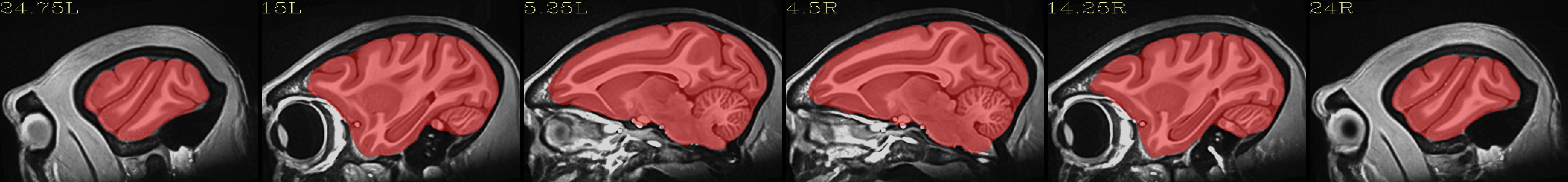
|
Skull-stripped NMT with 4-tissue segmentation |
|---|
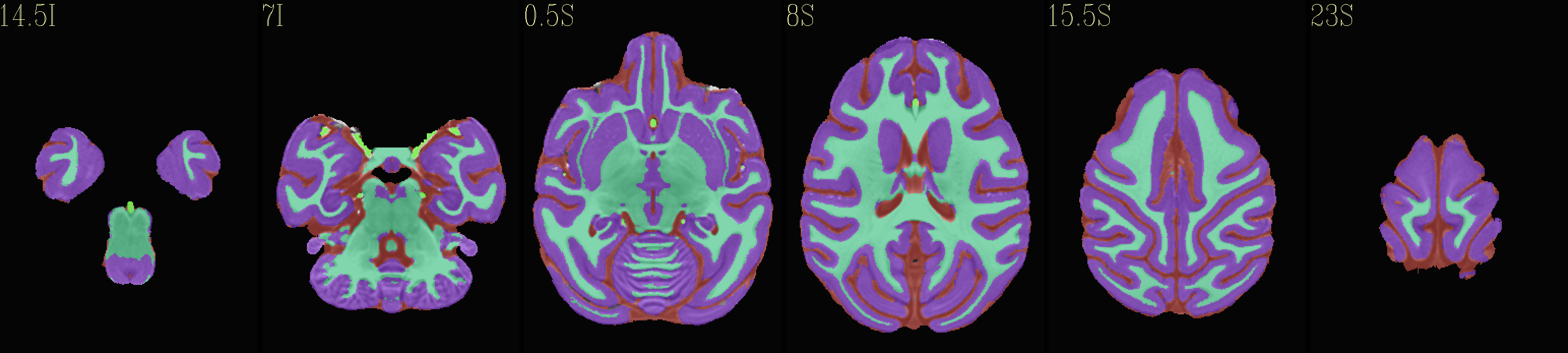
|
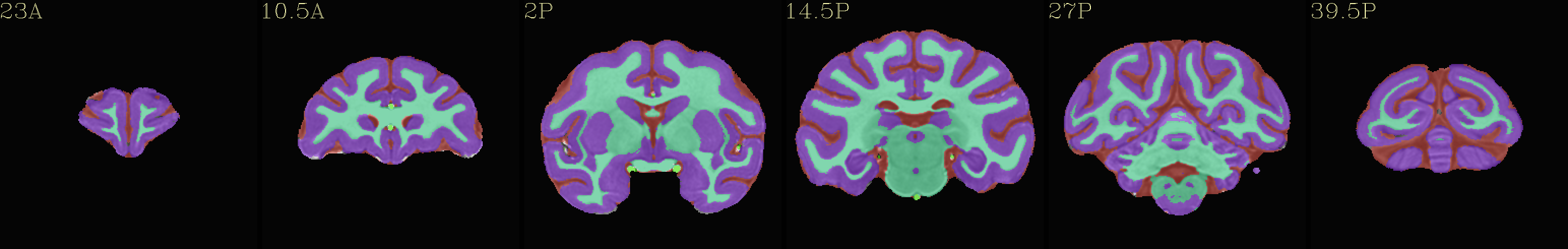
|
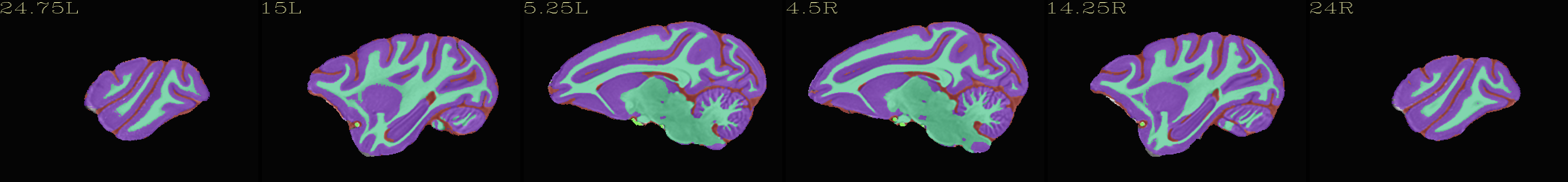
|
Skull-stripped NMT with D99 atlas |
|---|
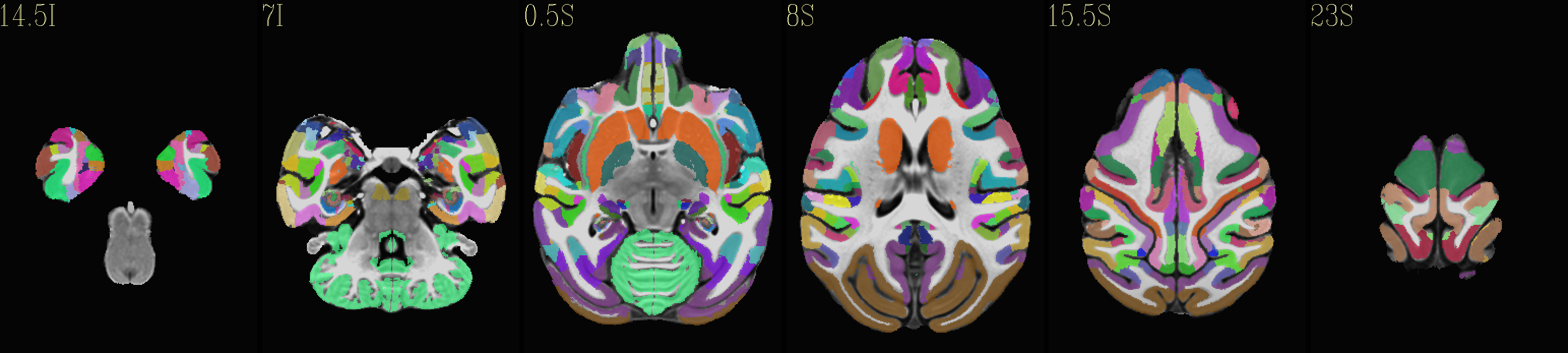
|
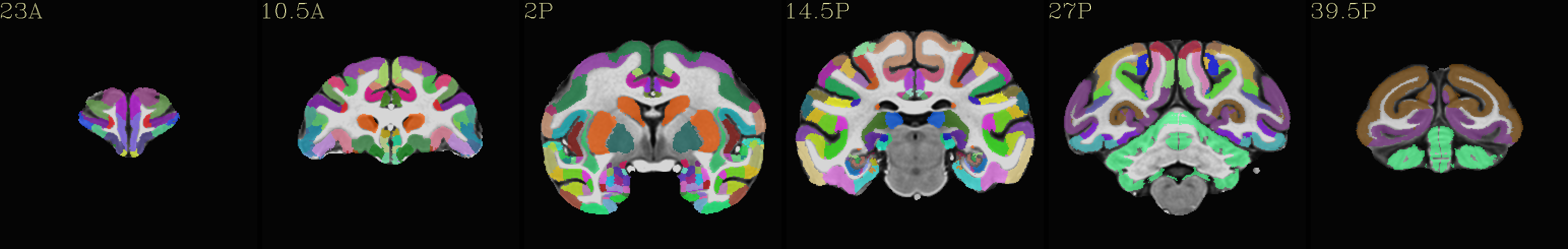
|
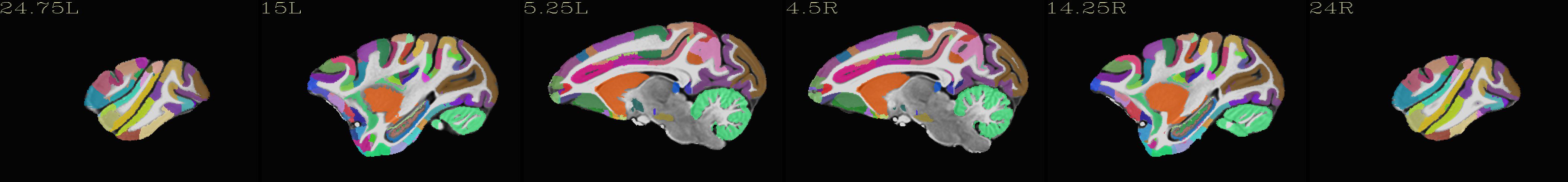
|
The script used to make these images with ``@chauffeur_afni`` is
here: do_view_nmt_v1.3.tcsh
Download¶
You can download and unpack the datasets in any of the following ways:
(the AFNI way) copy+paste:
@Install_NMT -nmt_ver 1.3
(the plain Linux-y terminal way) copy+paste:
curl -O https://afni.nimh.nih.gov/pub/dist/atlases/macaque/nmt/NMT_v1.3.tgz tar -xvf NMT_v1.3.tgz
- (the mouseclick+ way) click on this link,... and then unpack the zipped directory by either clicking on it or using the above
tarcommand.
Citation/questions¶
If you make use of the NMT v1.3 template or accompanying data in your research, please cite:
Seidlitz J, Sponheim C, Glen DR, Ye FQ, Saleem KS, Leopold DA, Ungerleider L, Messinger A (2018). “A Population MRI Brain Template and Analysis Tools for the Macaque.” NeuroImage 170: 121–31. https://doi.org/10.1016/j.neuroimage.2017.04.063.
Jung B, Taylor PA, Seidlitz PA, Sponheim C, Glen DR, Messinger A (2020). “A Comprehensive Macaque FMRI Pipeline and Hierarchical Atlas.” NeuroImage, submitted.
NMT v1.2¶
Contents¶
The following volumetric, surface and supplementary datasets are available as part of the download, but at present we would recommend using one of the more modern versions of the NMT for most applications.
atlases/ |
blood_vasculature.gii |
cerebellum.gii |
index.html |
lh.GM.gii |
lh.GM.inflated.surf.gii |
lh.mid.gii |
lh.mid.inflated.surf.gii |
lh.WM.gii |
lh.WM.inflated.surf.gii |
NMT_blood_vasculature_mask.nii.gz |
NMT_both.spec |
NMT_brainmask.nii.gz |
NMT_cerebellum_mask.nii.gz |
NMT.fullhead.nii.gz |
NMT_GM_cortical_mask.nii.gz |
NMT.nii.gz |
NMT_olfactory_bulb_mask.nii.gz |
NMT_segmentation_4class.nii.gz |
NMT_segmentation_CSF.nii.gz |
NMT_segmentation_GM.gz |
NMT_segmentation_WM.gz |
NMT_SS.nii.gz |
NMT_subject_align.csh |
NMT_subject_morph |
NMT_subject_process |
README.md |
rh.GM.gii |
rh.GM.inflated.surf.gii |
rh.mid.gii |
rh.mid.inflated.surf.gii |
rh.WM.gii |
rh.WM.inflated.surf.gii |
volumetric_transformations/ |
A SUMA view of some of the data in the NMT v1.2 |
|---|
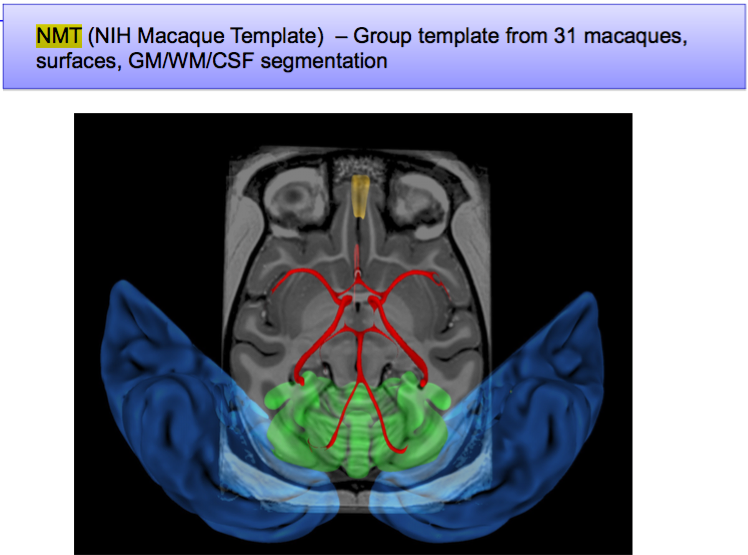
|
Download¶
You can download and unpack the datasets in any of the following ways:
(the AFNI way) copy+paste:
@Install_NMT -nmt_ver 1.2
(the plain Linux-y terminal way) copy+paste:
curl -O https://afni.nimh.nih.gov/pub/dist/atlases/macaque/nmt/NMT_v1.2.tgz tar -xvf NMT_v1.2.tgz
- (the mouseclick+ way) click on this link,... and then unpack the zipped directory by either clicking on it or using the above
tarcommand.
Citation/questions¶
If you make use of the NMT v1.2 template or accompanying data in your research, please cite:
Seidlitz J, Sponheim C, Glen DR, Ye FQ, Saleem KS, Leopold DA, Ungerleider L, Messinger A (2018). “A Population MRI Brain Template and Analysis Tools for the Macaque.” NeuroImage 170: 121–31. https://doi.org/10.1016/j.neuroimage.2017.04.063.
