10.3. @SSwarper base volumes¶
A brief description of the special multi-volume datasets used as a
reference for the program @SSwarper, which both skull-strips
and generates a nonlinear warp to a reference template space.
10.3.1. @SSwarper¶
For full details on this program, as well as how to integrate it
straightforwardly with afni_proc.py, please see @SSwarper’s
helpfile.
In short, this program iterates internally between skull-stripping and registration to a reference template brain to accomplish the dual goals of skull-stripping the input and estimating a nonlinear warp to a standard template space; these problems are not independent, and this program leverages that fact to provide reasonable results for both quite often.
While the final level of refinement (i.e., the patch size checked for
alignment) can be selected by the user, in general @SSwarper can
be pretty slow overall, of the order of a couple hours. However, the
biggest time-hog within it (3dQwarp) has been written using
OpenMP, so it can run on several CPUs simultaneously, saving time. If
you have the computer space, using something like 8-12 CPUs should
provide good speed up, greatly reducing the overall run time.
The primary final outputs are:
A skull-stripped version of the input anatomical, in its original space
A set of transformations that can be concatenated to transform from the original anatomical dataset’s space (“source”) to the grid of the reference template (“base”)
Fancy-schmancy JPG image files showing the alignment of the features of the data sets, namely an edge-ified version of one over the other (this facilitates the quality check of the alignment and skull-stripping– don’t forget to look at your analyzed data!!).
10.3.2. Reference templates for @SSwarper¶
In order to use @SSwarper, the user needs to provide a
specially-created, multi-volume dataset as a reference. It has five
volumes to it, and (reading from the program’s help), these are:
[0] = skull-stripped template brain volume
[1] = skull-on template brain volume
[2] = weight mask for nonlinear registration, with the
brain given greater weight than the skull
[3] = binary mask for the brain
[4] = binary mask for gray matter plus some CSF (slightly dilated)
-- this volume is not used in this script
-- it is intended for use in restricting FMRI analyses
to the 'interesting' parts of the brain
-- this mask should be resampled to your EPI spatial
resolution (see program 3dfractionize), and then
combined with a mask from your experiment reflecting
your EPI brain coverage (see program 3dmask_tool).
We now distribute such volumes for a growing number of spaces. They can typically be identified by having “_SSW” attached after their prefix, such as:
MNI152_2009_template_SSW.nii.gz (the original!)
HaskinsPeds_NL_template1.0_SSW.nii.gz: the Haskins Pediatric (HP) template
The above sub-volumes need to be present in the -base .. dataset
for @SSwarper, in the correct order. You are welcome to make your
own. Commented scripts are provided below for demonstrating how some
of the above examples were made, to give you some help along the way.
MNI152_2009_template_SSW.nii.gz (5 volumes; one volume per column; axi, cor, sag views) |
|---|
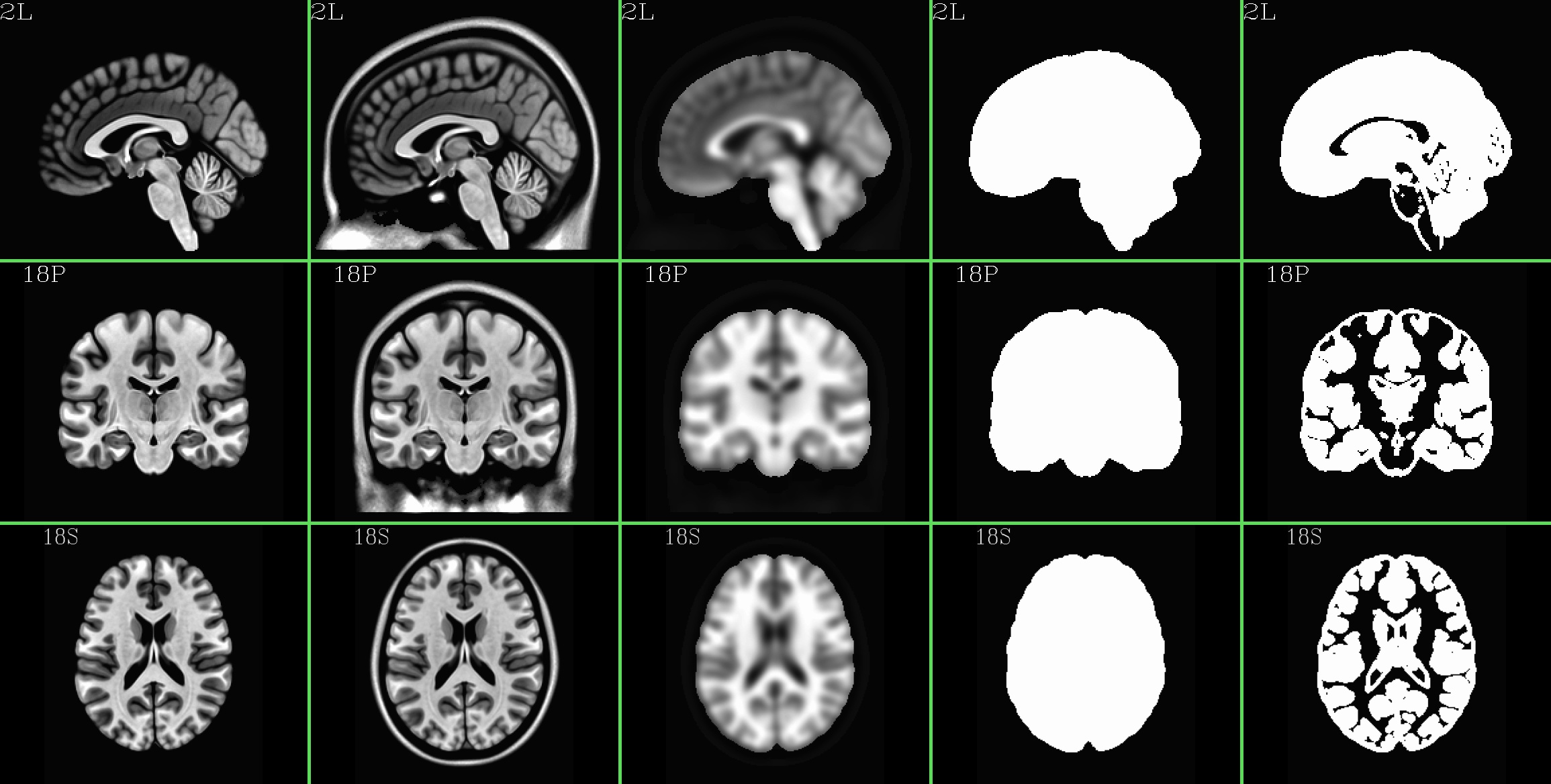
|
TT_N27_SSW.nii.gz (5 volumes; one volume per column; axi, cor, sag views) |
|---|
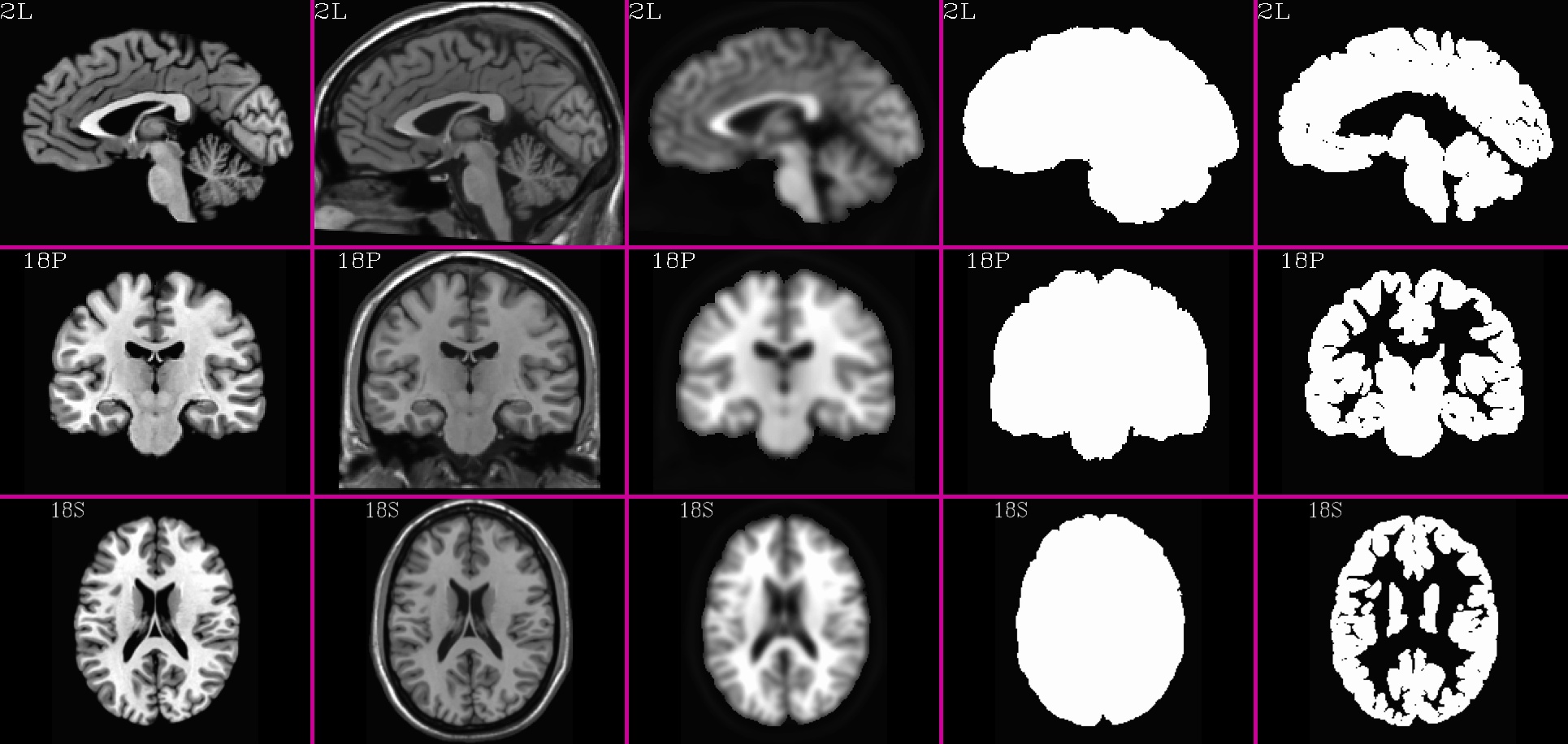
|
HaskinsPeds_NL_template1.0_SSW.nii.gz (5 volumes; one volume per column; axi, cor, sag views) |
|---|
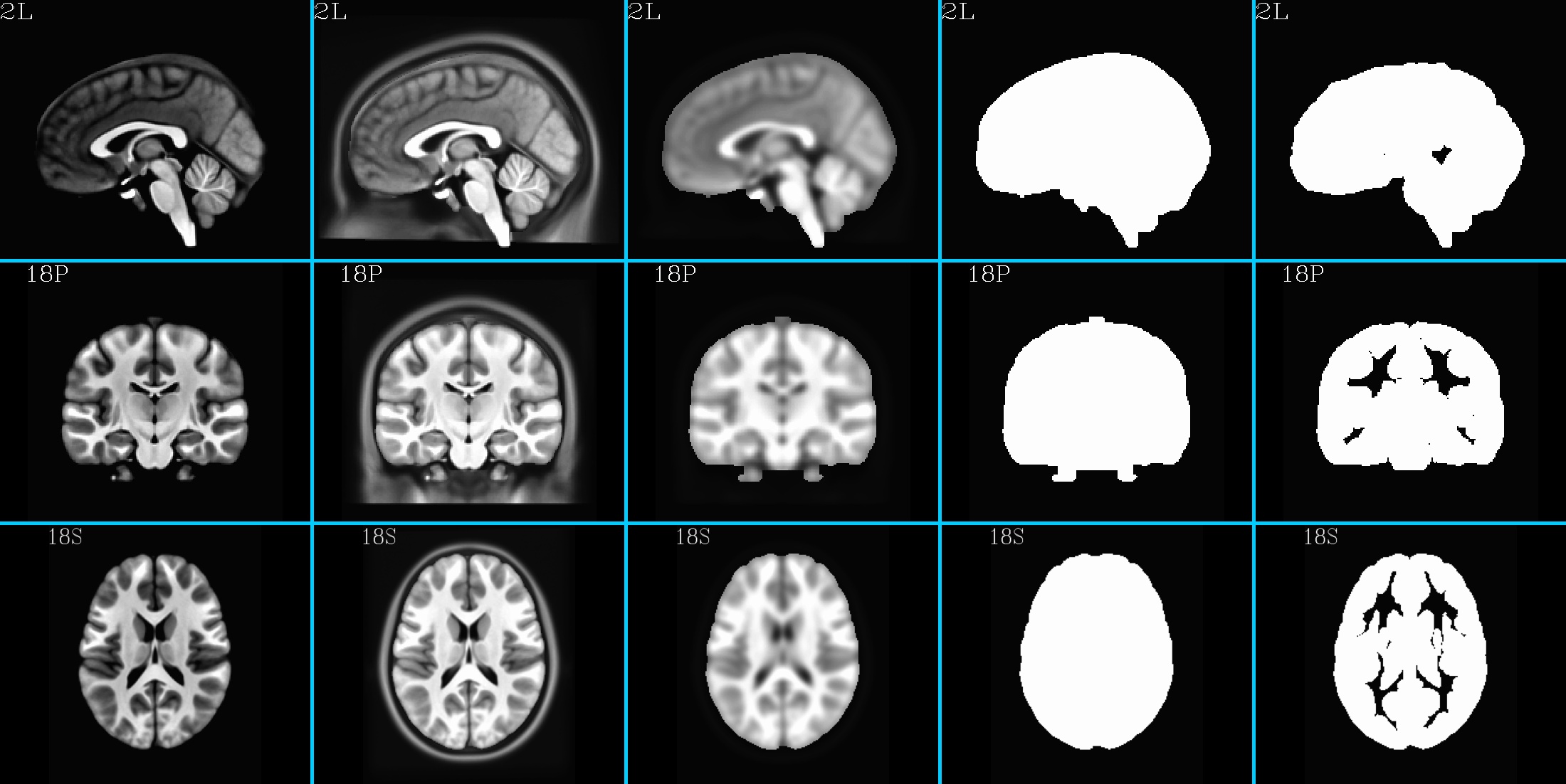
|
10.3.3. Example for making a reference dset: TT_N27_SSW.nii.gz¶
Here are some example scripts for making each of the five volumes of
the TT_N27_SSW.nii.gz dset (do_TT_0[0-4]_*tcsh) and then for
concatenating them all (do_TT_05_*tcsh). Each script is commented.
Note that the making of the “skull-on” [1] volume is a bit of a special case here for TT_N27_SSW.nii.gz: the version of this brain with a skull on was not in the same space as the skull-stripped [0] volume, and therefore it had to be brought into it using parameters stored in the header file using “adwarp”. For most scenarios, this step would not need to be done as such (it would usually just be copied, for example).
Scripts for making: TT_N27_SSW.nii.gz |
Description |
|---|---|
Make the “skullstripped” [0] volume. Basically, just copies a skull-stripped reference volume. |
|
Make the “skull-on” [1] volume. See the text above for why this is such an unusually involved step here. |
|
Make the blurry volume that includes a dimmed skull, as the [2] volume. This is done by using the already-made volumes [0] and [1]. |
|
Make the whole brain mask [3] volume. |
|
Make the (inflated, or “generous”) gray matter tissue mask [4] volume. |
|
Concatenate all the individual bricks into a single, multi-volume masterpiece. |
10.3.4. Example for making a reference dset: HaskinsPeds_NL_template1.0_SSW.nii.gz¶
Here are some example scripts for making each of the five volumes of
the HaskinsPeds_NL_template1.0_SSW.nii.gz dset
(do_HP_0[0-4]_*tcsh) and then for concatenating them all
(do_HP_05_*tcsh). Each script is commented.
Here as well, the making of the “skull-on” [1] volume is a bit of a special case here: there was no prior volume with a skull for this data set. Therefore, we “borrowed” the skull from that of another reference template; we performed linear-affine alignment to the skull-bearing “mni_icbm152_t1_tal_nlin_sym_09a.nii” volume (freely available for download under the “ICBM 2009a Nonlinear Symmetric 1×1x1mm template” section from here), and applied
Also, the final volume’s inflated gray matter (GM) map in volume [4] was made starting from the HP template’s associated atlas (“HaskinsPeds_NL_atlas1.0+tlrc”).
Scripts for making: TT_N27_SSW.nii.gz |
Description |
|---|---|
Make the “skullstripped” [0] volume. Basically, just copies a skull-stripped reference volume. |
|
Make the “skull-on” [1] volume. See the text above for why this is such an unusually involved step here. |
|
Make the blurry volume that includes a dimmed skull, as the [2] volume. This is done by using the already-made volumes [0] and [1]. |
|
Make the whole brain mask [3] volume. |
|
Make the (inflated, or “generous”) gray matter tissue mask [4] volume; uses the associated atlas to define a GM map, which gets inflated. |
|
Concatenate all the individual bricks into a single, multi-volume masterpiece. |
