12.5.9. Post-preproc, III: ROI map preparation¶
Overview¶
The FreeSurfer (FS) defined regions may be useful for ROI-based
analysis or for tracking. However, it should be noted that among the
GM regions can be non-local bits of general cortex that were not
specifically classified. These come under the FS-labelling of
“Left-Cerebral-Cortex” (FS #3 and AFNI REN #2) and
“Right-Cerebral-Cortex” (FS #42 and AFNI REN #22) in both the “2000”
and “2009” parcellations. These leftover-ish voxels don’t really form
a solid region unto itself in the sense that most people would likely
want for any ROI-based analysis, even though they may indeed be in the
GM tissue mask. So, in order to prepare for analyses where one just
wants localized regions, we demonstrate de-selecting these regions
from the GM maps in DT space that were created using
fat_proc_map_to_dti in the step, “Post-preproc, II: mapping ROIs to DTI space”.
Additionally, specifically in preparation for tracking, one might want to inflate the GM regions, which we consider “targets” for the tracking function, in a controlled way:
they can extend up to, but not too much into, DTI-defined WM (which, for healthy adult humans, is usually determined by where FA>0.2, in a standard proxy)
they can extend up to, but not into, any other GM regions
The regions can be inflated with these properties by the AFNI-FATCAT
function 3dROIMaker, which is described in more detail (including
other options for controlling the ROIs) HERE.
ROI selection and 3dROIMaker¶
Note
At some point there may be a separate function created for combining these capabilities. For the present, we just describe one example of how the GM regions can be selected and then inflated.
Proc: We demonstrate de-selecting the non-clustered regions that may (or may not, depending on FS) be in the GM parc/seg map. This pretty much depends on knowing what one wants to get rid of, by ROI number designation. The labeltable from the original file is reattached.
Finally, 3dROIMaker is run to controlledly inflate the regions
that are left, with respect to the FA data (if one had non-adult or
non-human data, one might reconsider the FA-derived proxy for defining
where WM is). NB: I think I might most prefer using the
-skel_stop_strict option to constrain the inflation. The resulting
set of commands are here:
# for I/O, same path variable as above
set path_P_ss = data_proc/SUBJ_001
# for variable simplicity, just go to the dir with the ROI maps
cd $path_P_ss/dwi_05
# list all (= 2, probably) renumbered GM maps
set ren_gm = `ls indt_*_REN_gm.nii.gz`
# supplementary files used in ROI process: FA map and WB mask
set wmfa = "dt_FA.nii.gz"
set wbmask = "dwi_mask.nii.gz"
# process each GM map
foreach gm_in ( $ren_gm )
set gm_sel = "sel_${gm_in}"
# apply the ROI (de)selector
3dcalc \
-echo_edu \
-a $gm_in \
-expr 'a*not(equals(a,2)+equals(a,22))' \
-prefix $gm_sel
# keep the labeltables from the original
3drefit \
-copytables \
$gm_in \
$gm_sel
set gm_roi = "${gm_sel:gas/.nii.gz//}"
# inflate GM ROIs up to (but not overlapping) the FA-WM
3dROIMaker \
-inset $gm_sel \
-refset $gm_sel \
-skel_stop_strict \
-skel_thr 0.2 \
-wm_skel $wmfa \
-inflate 1 \
-neigh_upto_vert \
-mask $wbmask \
-prefix $gm_roi \
-nifti
end
-> produces output in the same directory ‘data_proc/SUBJ_001/dwi_05/’, since all volume files occupy the same space/grid:
Directory structure for example data set |
|---|
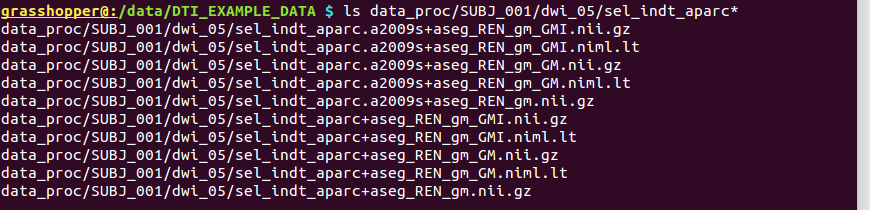
|
Output files made to select only clumpy GM ROIs and then to perform controlled inflation. |
Outputs of |
the above |
|---|---|
sel_indt_aparc*+aseg_REN_gm.nii.gz |
volumetric NIFTI file, 3D; the GM region map without the deselected ROIs. The same labeltable from the original input is contained within the file’s header. |
sel_indt_aparc*+aseg_REN_gm_GMI.nii.gz |
volumetric NIFTI file, 3D; the output of |
sel_indt_aparc*+aseg_REN_gm_GMI.niml.lt |
text file; the labeltable of the NIFTI file with the same root name. |
sel_indt_aparc*+aseg_REN_gm_GM.nii.gz |
volumetric NIFTI file, 3D; the output of |
sel_indt_aparc*+aseg_REN_gm_GM.niml.lt |
text file; the labeltable of the NIFTI file with the same root name. |
To view the dual points of 1) inflating the GM ROIs and 2) constraining that inflation, we show images of before-and-after inflation, for both the “2000” and “2009” parcellations. The b/w ulay is the binary mask where FA>0.2, representing the DTI-based proxy for WM (and within which tracking normally occurs for healthy adult humans). Note that in the pre-inflation cases, one can often see GM ROIs following the contours of the FA-WM, but there might be slight gaps due to either transformation, partial voluming, etc. Such regions might create artificial “misses” in the tracts, which don’t leave the FA>0.2 boundaries to reach the GM they (possibly) should. Conversely, in cases where the GM follows the FA-WM boundary well, we wouldn’t want inflation pouring out into the WM unnecessarily.
Note
When viewing the following montages, it might make sense to open corresponding montages of the inflated and non-inflated maps in browser tabs and then toggling views between them– that should highlight both of the main points.
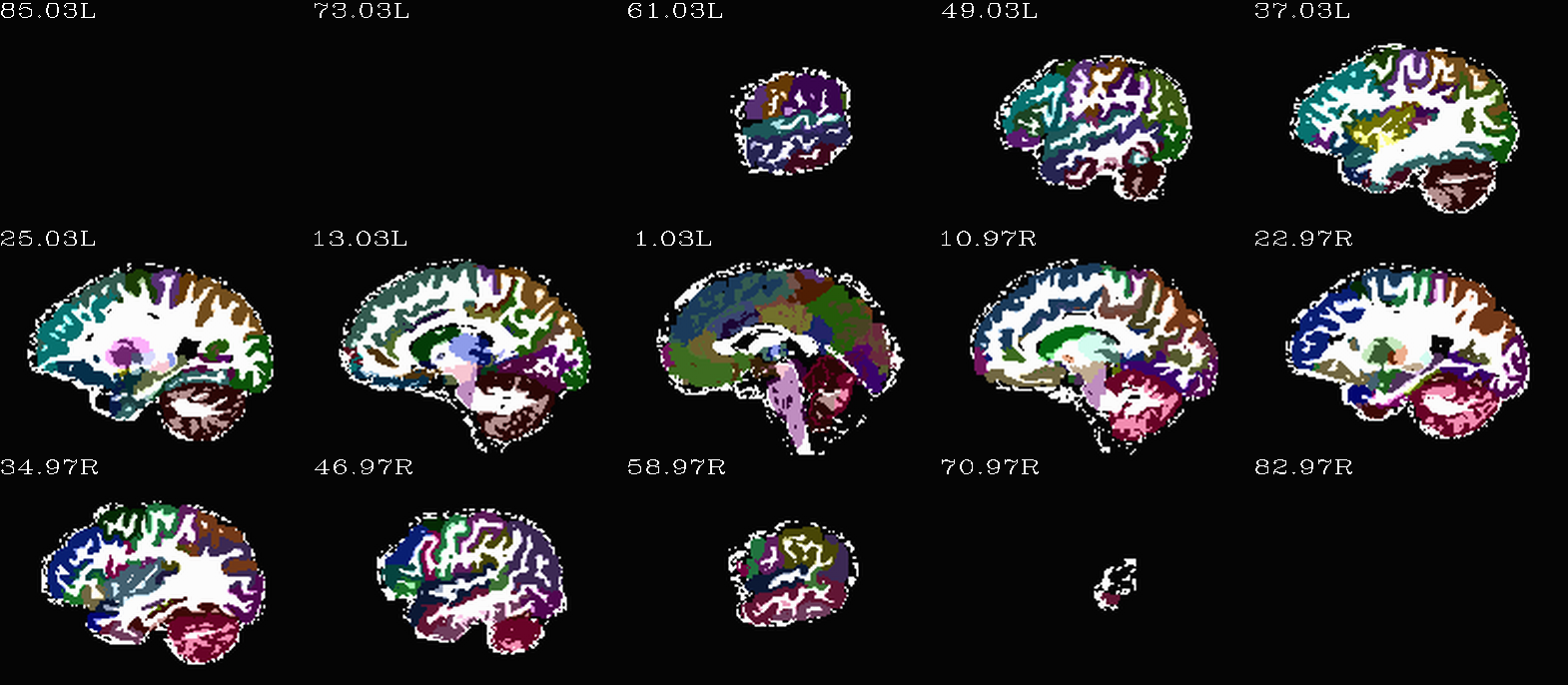
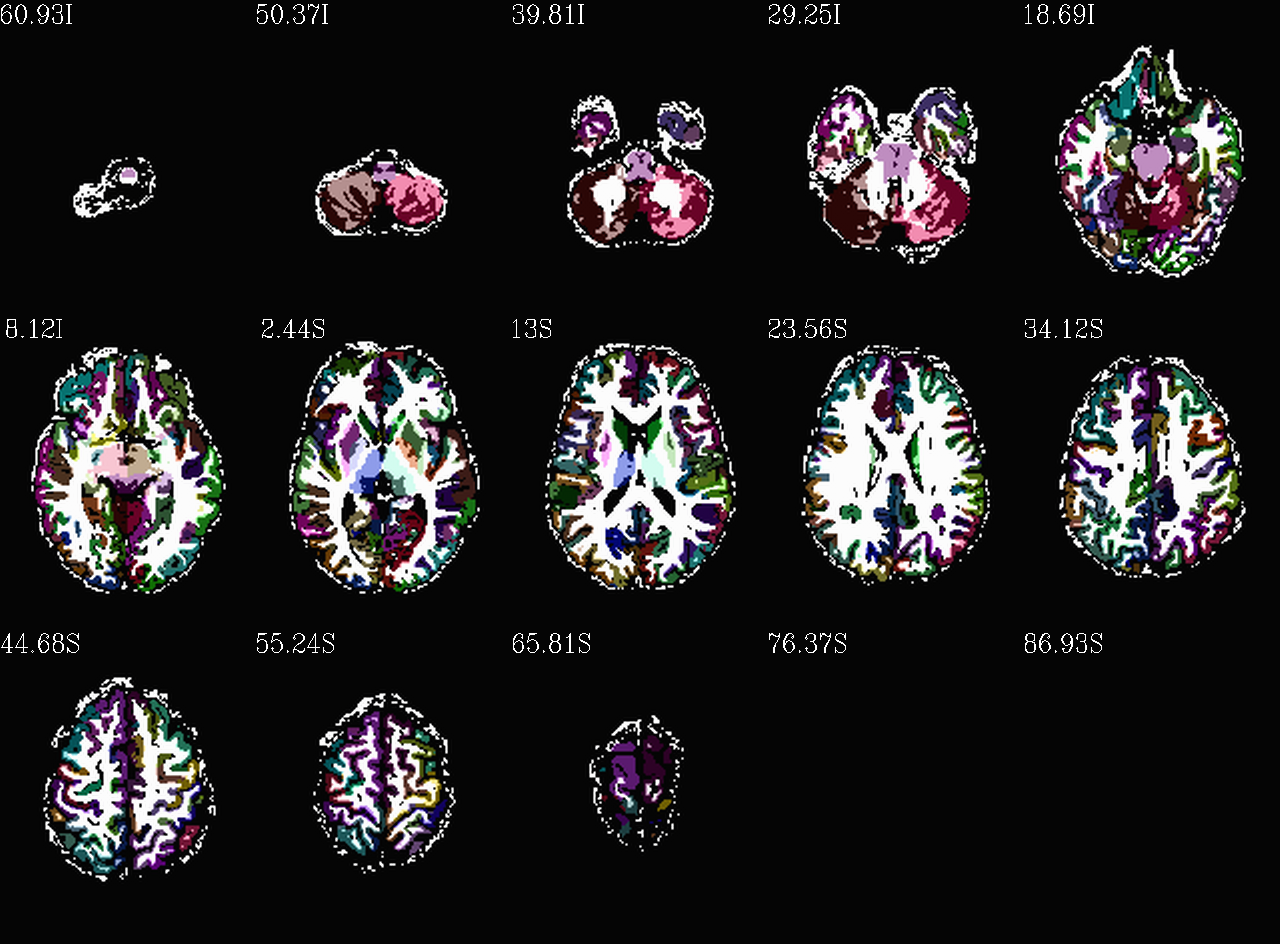
Images comparing the “2000” inflated and non-inflated GM maps |
(just axi and sag views) |
|---|---|
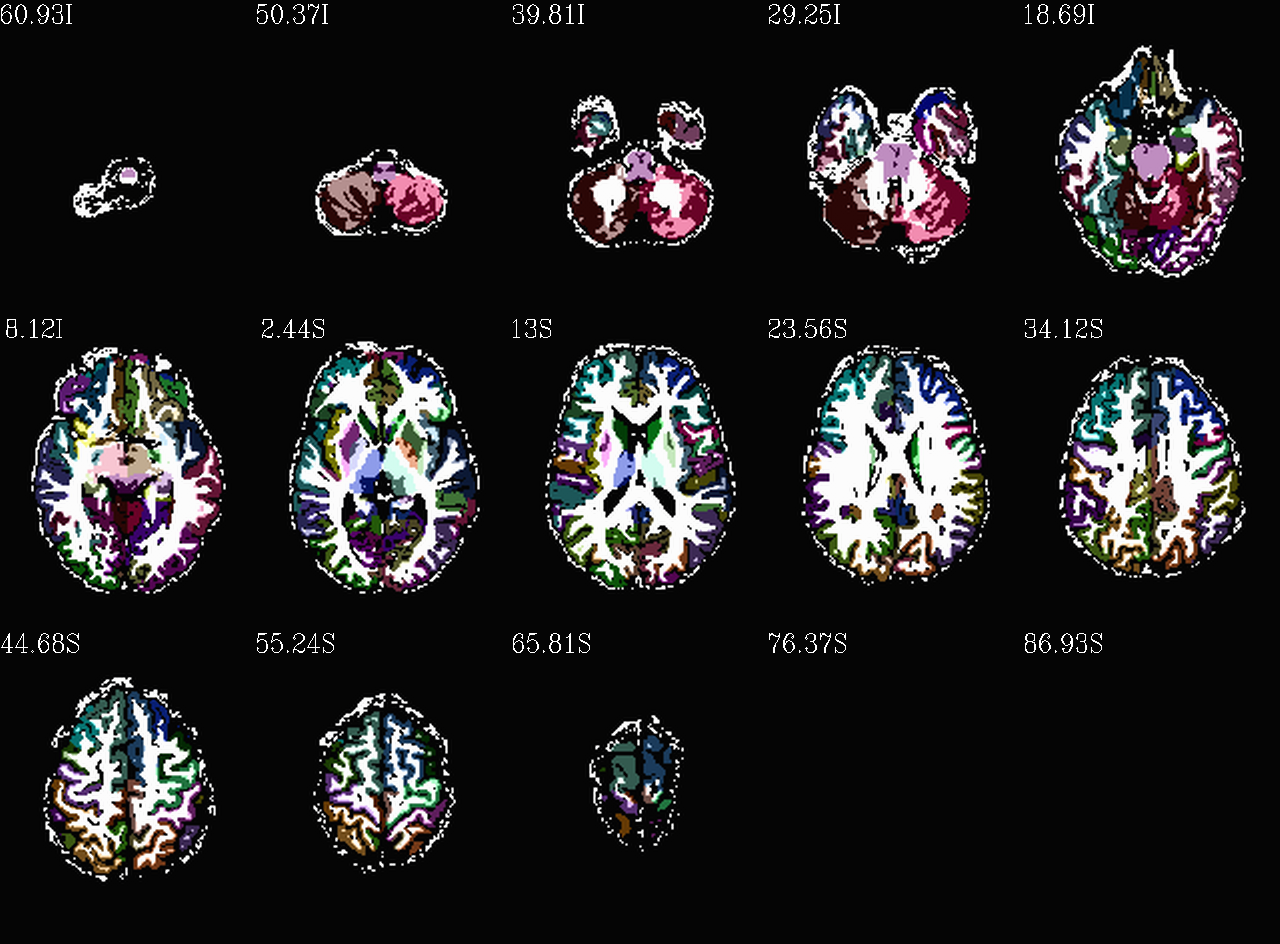
|
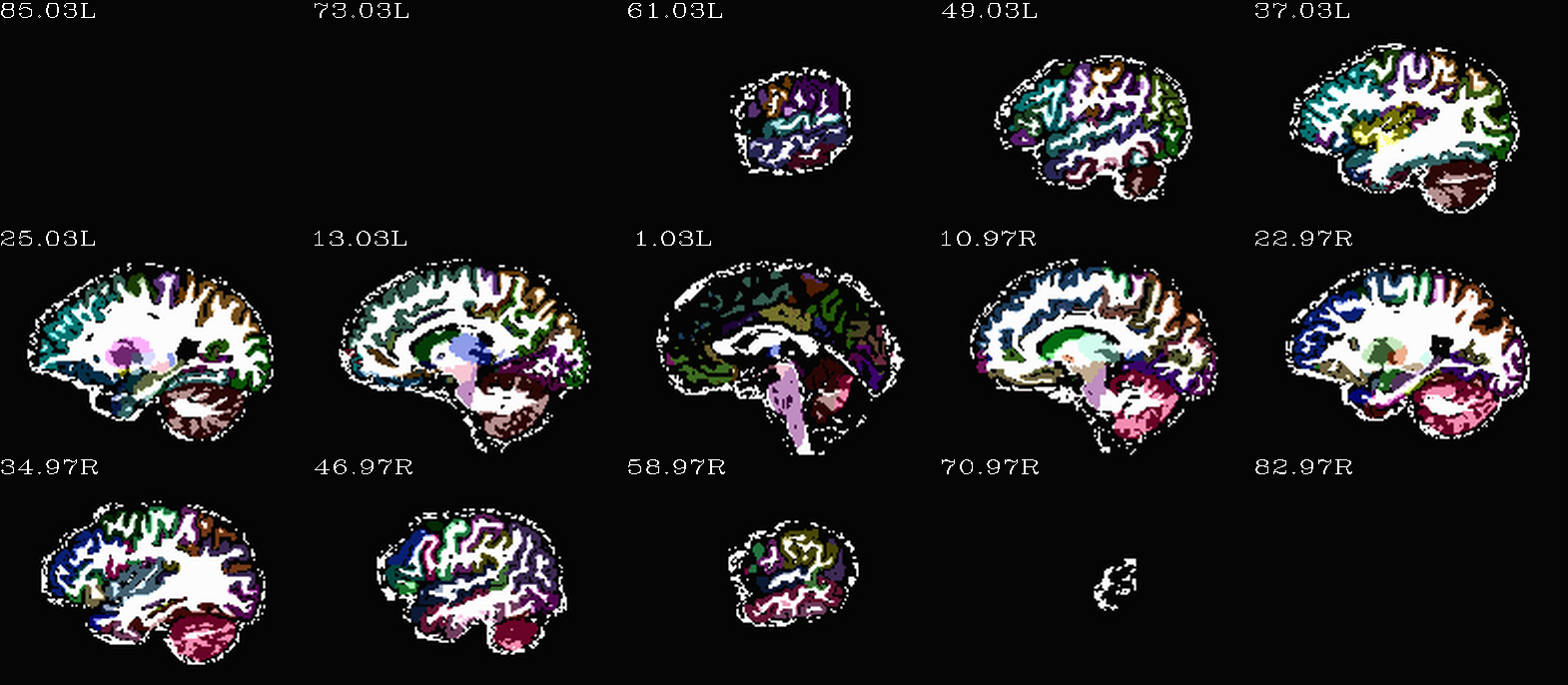
|
Non-inflated “2000” parc/seg map (after the non-regional ROIs were removed) olayed on FA>0.2 binary map ulay. |
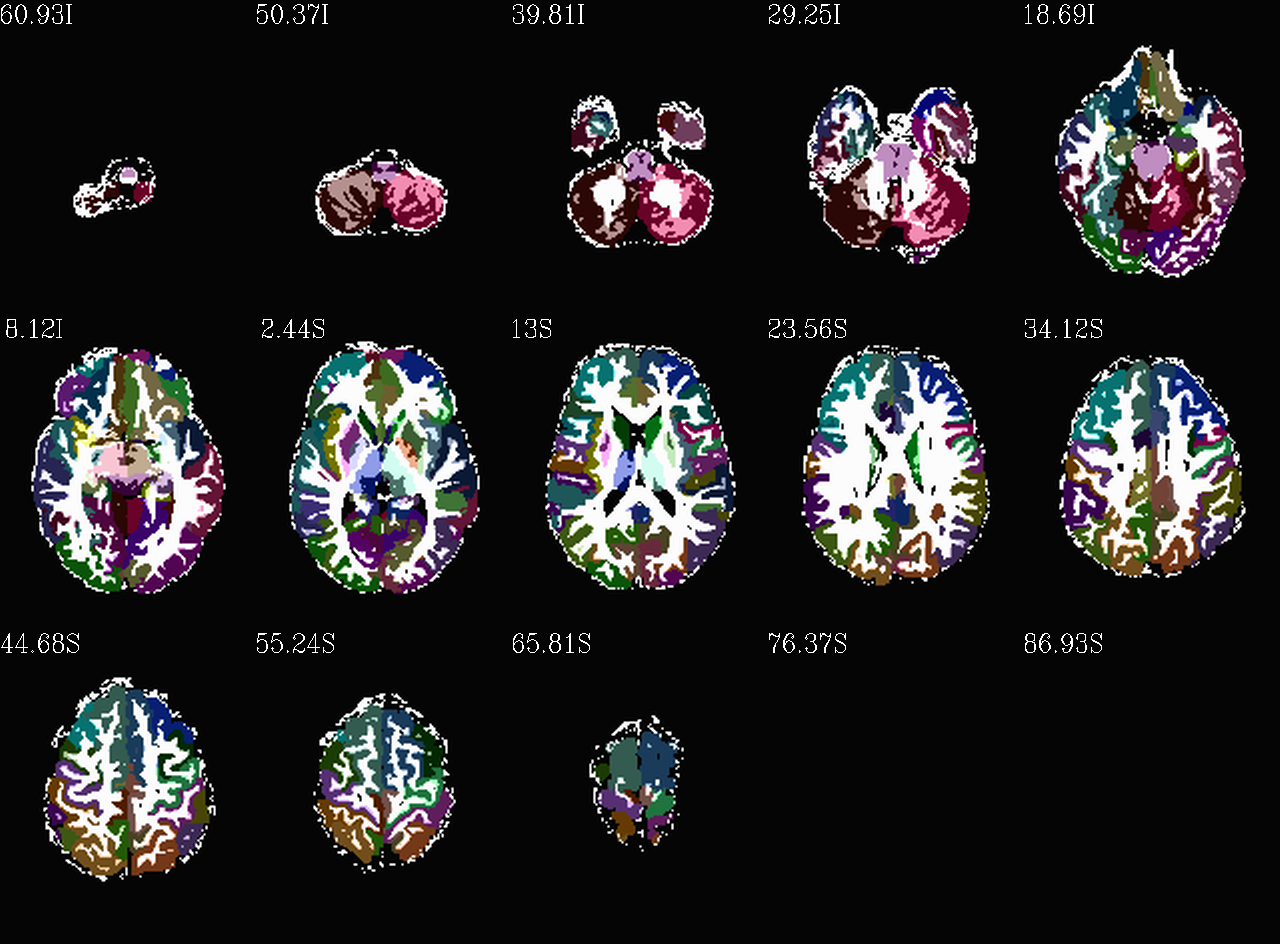
|
|
Inflated “2000” parc/seg map (after the non-regional ROIs were removed) olayed on FA>0.2 binary map ulay. |
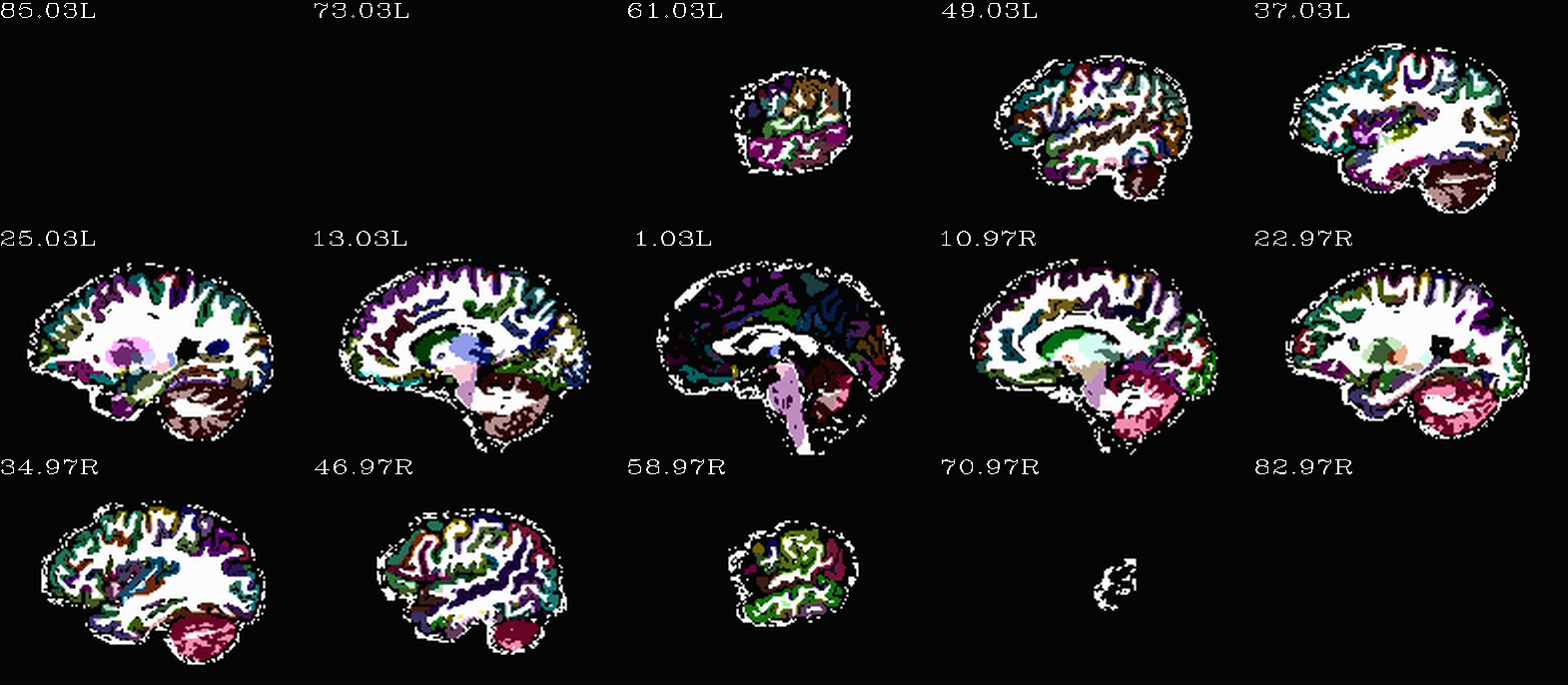
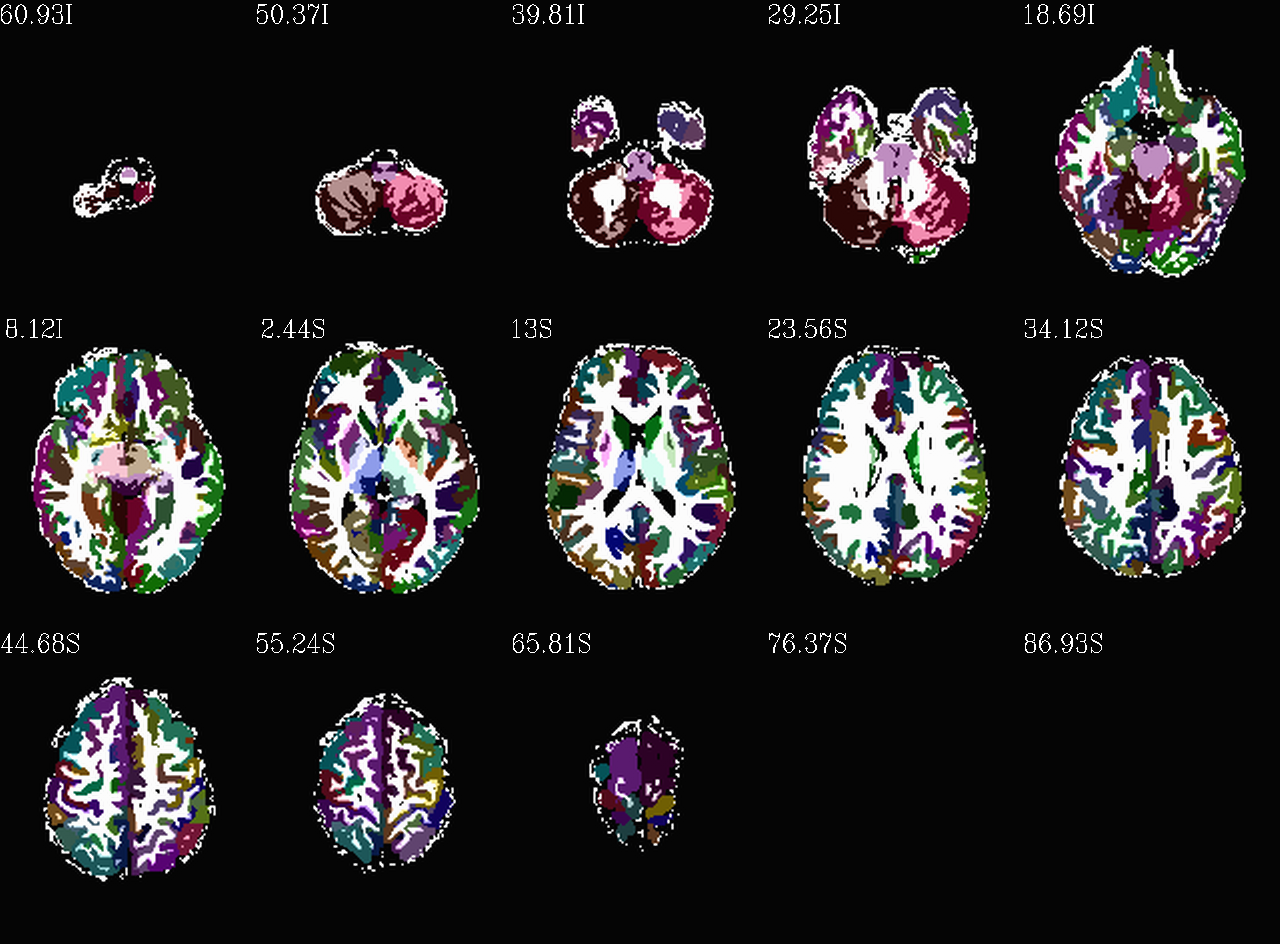
Images comparing the “2009” inflated and non-inflated GM maps |
(just axi and sag views) |
|---|---|
|
|
Non-inflated “2009” parc/seg map (after the non-regional ROIs were removed) olayed on FA>0.2 binary map ulay. |
|
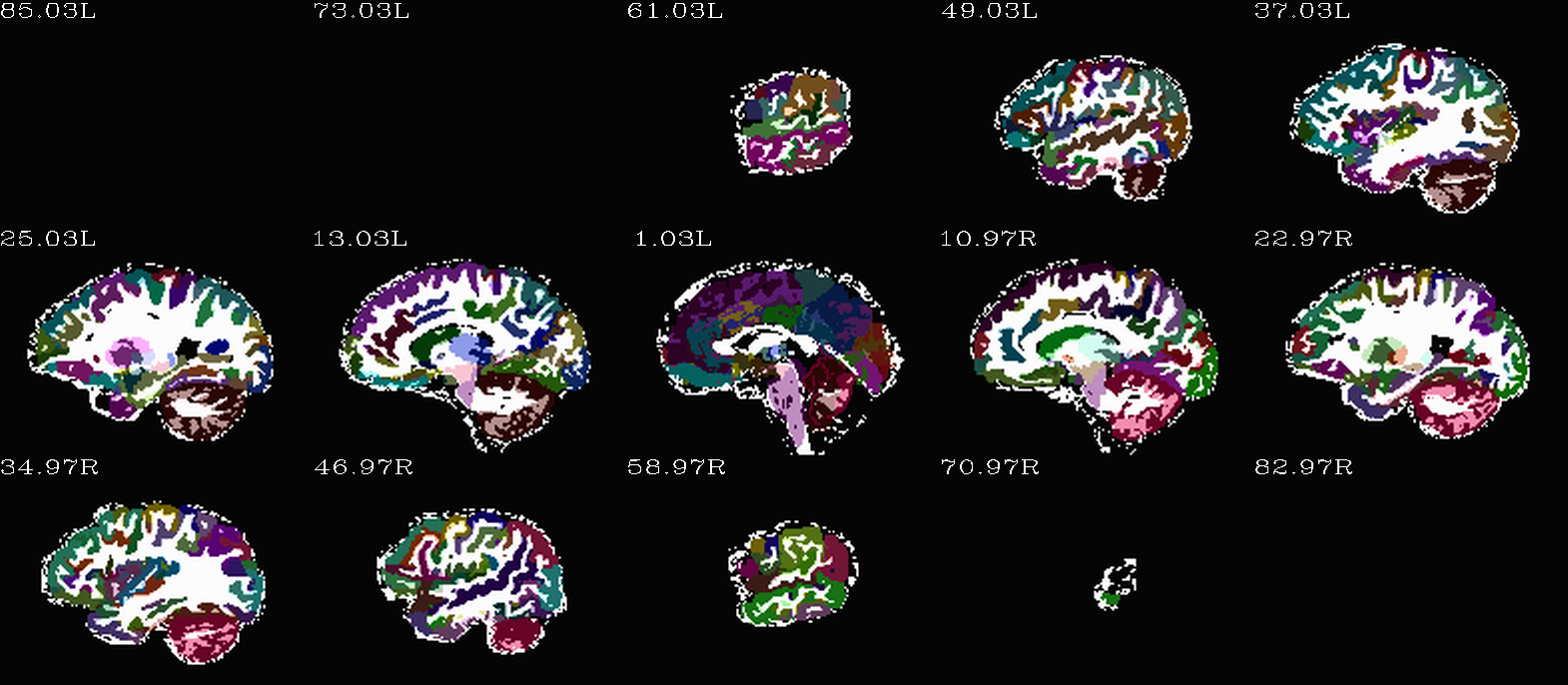
|
Inflated “2009” parc/seg map (after the non-regional ROIs were removed) olayed on FA>0.2 binary map ulay. |
