12.5.4. Pre-preproc, II: examine and filter¶
Overview¶
Sometimes volumes are so corrupted by distortion (lots of dropout,
subject motion, etc.) it makes sense to remove it from the
analysis. It has to be removed from both the list of
gradient/b-matrices, as well as from the volume set. Additionally, if
matching AP and PA (blip up/down) pairs of dsets have been acquired,
then the same volumes need to be removed from both sets. The
fat_proc_select_vols provides a wrapper for a straightforward,
user-interactive GUI (thanks, Justin Rajendra!) to be able to:
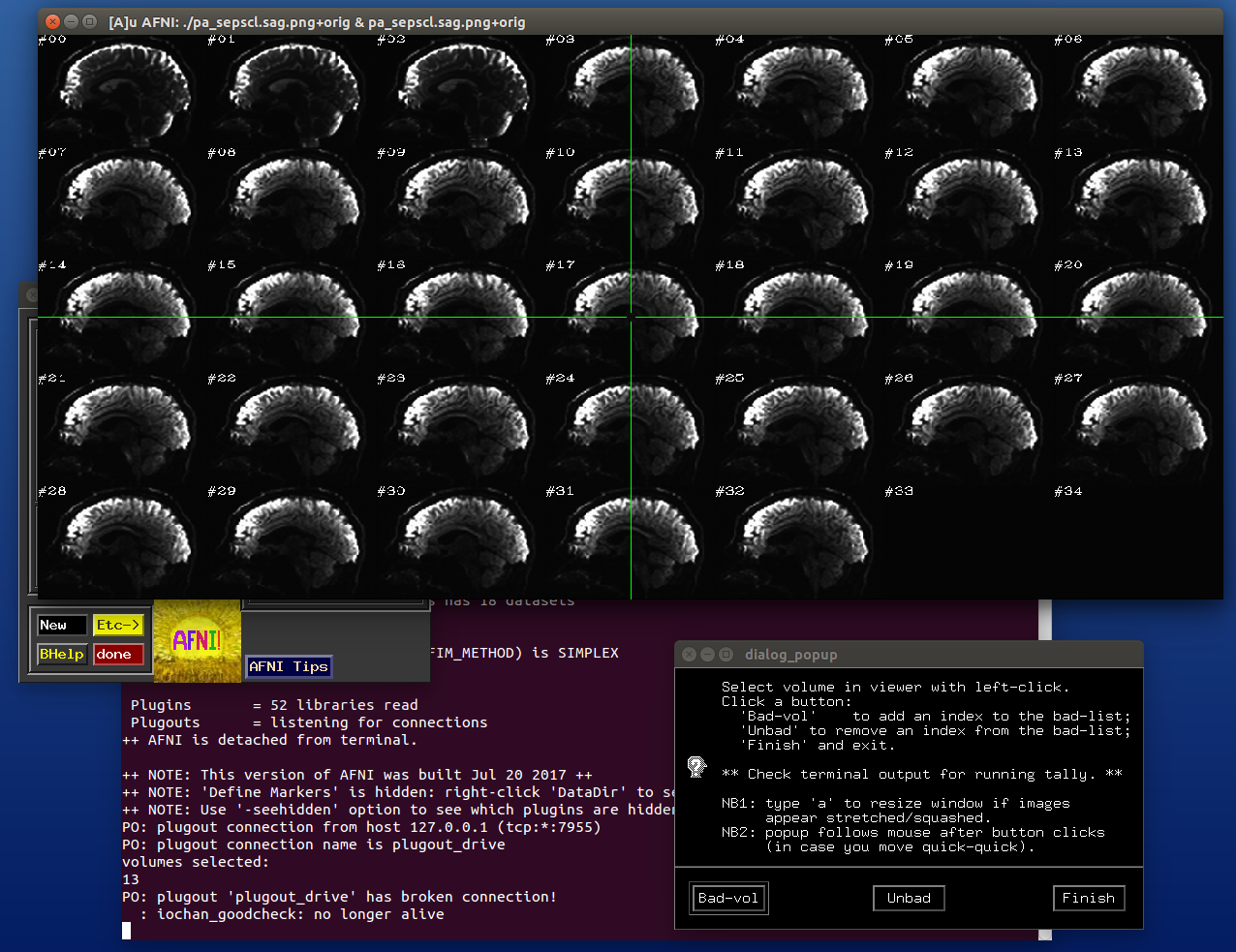
view a slice of each DWI in a dset simultaneously and to click on it to add it to a ‘bad’ list that is stored in a text file;
use multiple dsets to add to the list, so one can easily define a single list that is the union of ‘bad’ volumes;
create an AFNI-formatted string for subbrick/list selection that can be applied to both volumes/gradient files;
and keep a text-file record the applied filter, for future reference.
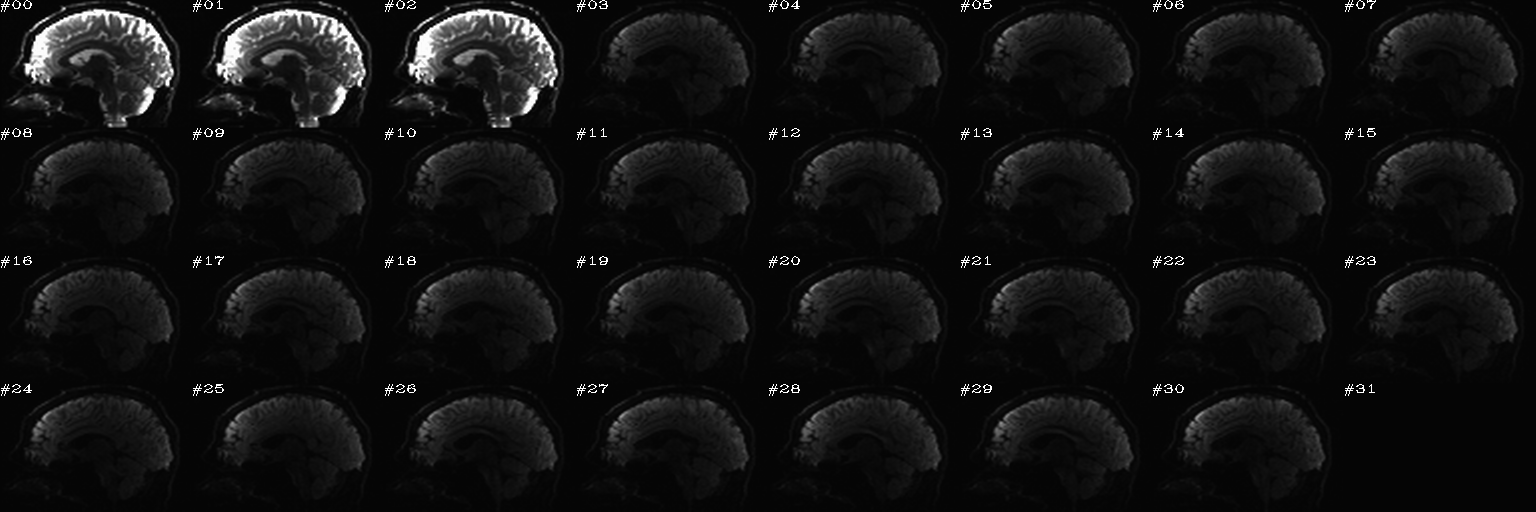
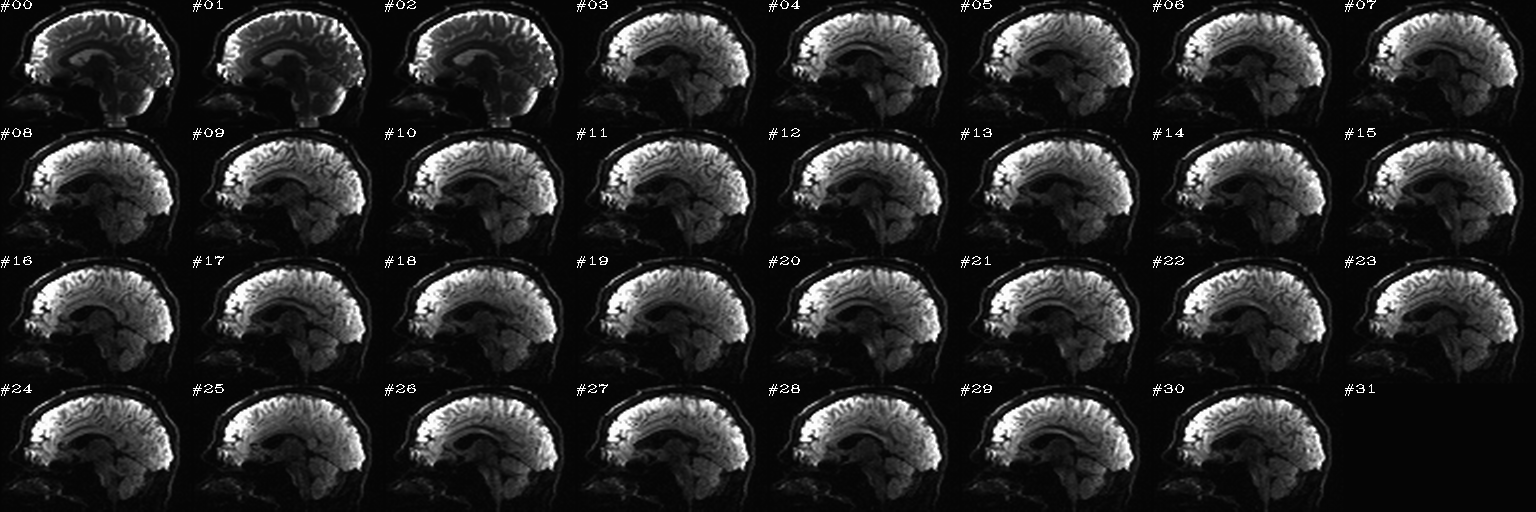
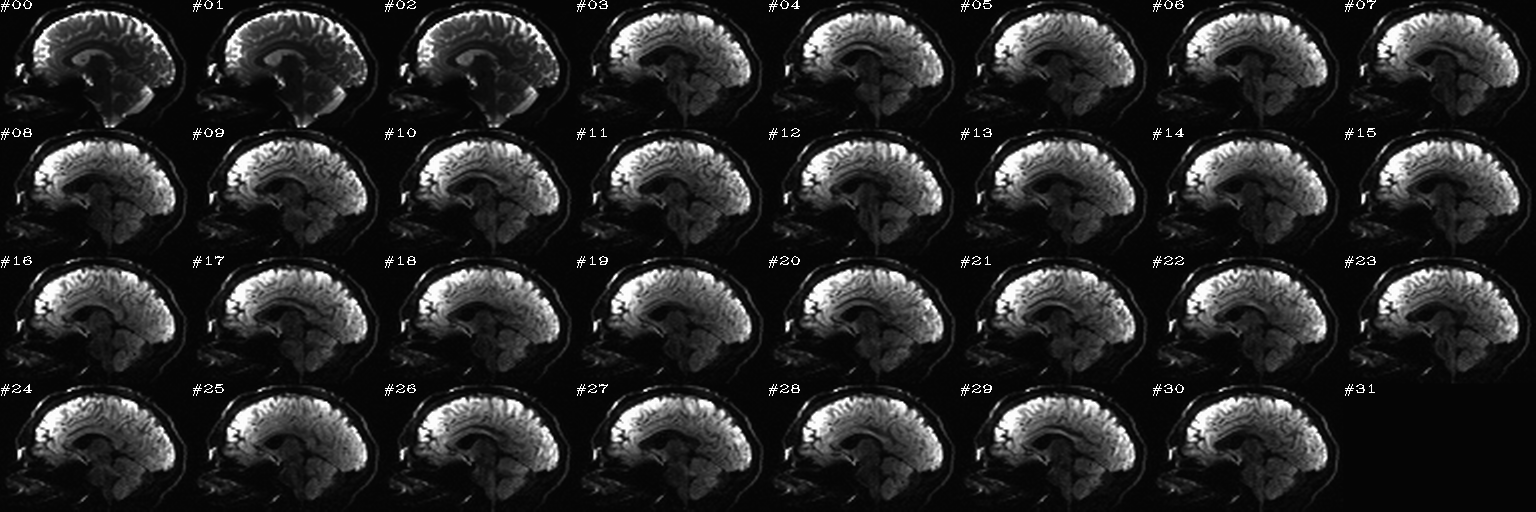
For slice viewing, I generally prefer using the sagittal view with separate scaling per volume. Generally, the presence of one or more bad slices is most readily apparent in these, IMHO.
The purpose of fat_proc_select_vols is to allow a user to
select bad volumes that should be removed, and to automatically
generate a list of good volumes to keep. The tool works by
selecting bad volumes because we assume that, in any given study,
there will be fewer bad volumes than good ones– if this is not the
case, your data+study might be in big trouble!
In AFNI, one can use “sub-brick” selectors to choose subsets of a data
set to input into a function. For example, let’s say AAA.nii.gz is a
4D data set containing 20 volumes. If you wanted to keep only the
first ten, then you would input AAA.nii.gz'[0..9]'; if you wanted
the last ten, this could be either AAA.nii.gz'[10..19]' or
AAA.nii.gz'[10..$]', as the “$” is a special character meaning “to
the end of the list”; if you wanted only volumes #0-5, 8 and 16-18,
you would input AAA.nii.gz'[0..5,8,16..18]'; etc. These selectors
can also be applied to text files, selecting indices of either rows or
columns using BBB.txt'[0..9]' or BBB'{0..9}', etc. See the
help of 3dcalc for more information.
Thus, the primary output of fat_proc_select_vols is a file
containing the selector string part of those above commands, plain and
simple. For example, in the example performed below, the whole
rigmarole with fat_proc_select_vols is to produce of file called
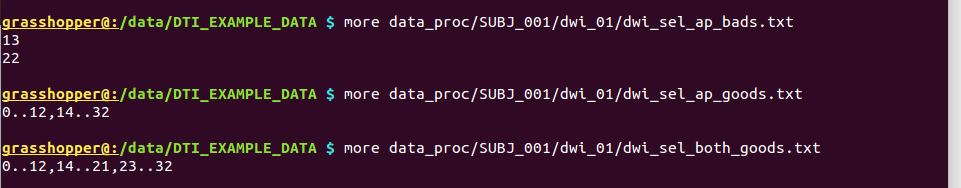
“dwi_sel_both_goods.txt” that contains a single line:
0..12,14..21,23..32
... which is the string selector of good volumes to keep from each of the AP and PA dsets. Note that that kind of simply structured file could be made in many different ways– feel free to use other methods. We do find value in A) users looking at and inspecting their own data for artifacts and B) having a written record of what they decided to filter.
Note
There are no brackets in the desired selector string of
good volumes. This is because the user may have either
row or column data of gradient and b-value information,
which would require different brackets. We deal with that
hassle internally in the fat_proc_filter_dwis function
so that the user doesn’t have to. You’re welcome.
The fat_proc_filter_dwis function applies the AFNI-formatted
selector string from fat_proc_select_vols (or, from any source,
actually) to input volume and gradient files. Yup, that’s it.
fat_proc_select_vols: GUI to select baddies (and make good list)¶
Note
In this example dset, there aren’t actually obviously bad volumes. One would normally look for signs of data corrupted by within-TR subject motion, etc. Here, “bad” volumes are just selected for the sake of didacticism– I hope you’re happy.
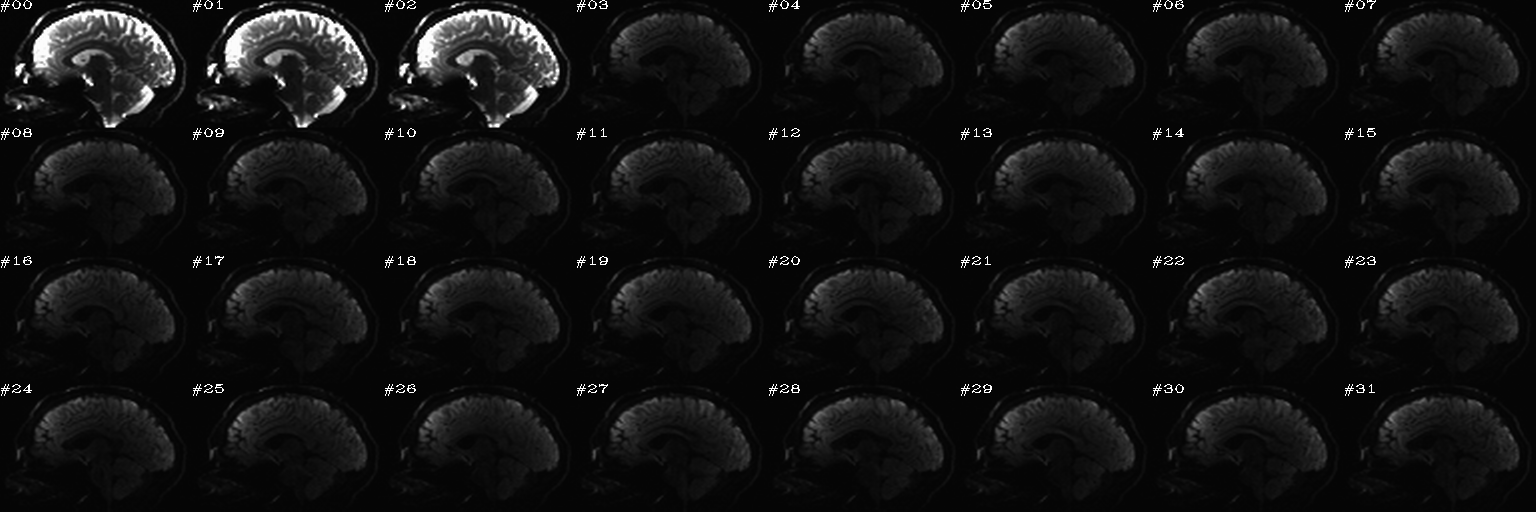
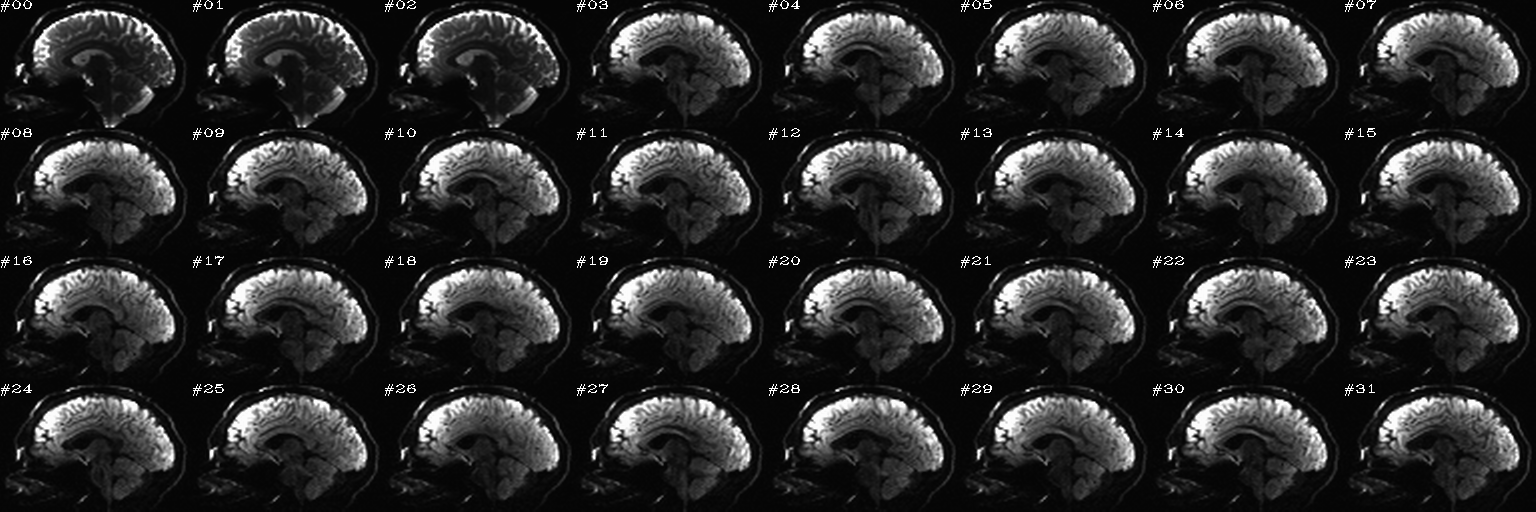
Proc: A paired set of N DWIs with opposite phase encode directions (AP and PA); the following opens up an earlier-made set of sagittal views of each volume, with brightness scaled separately per slice, and one can click on bad volumes to add them to the list; then, in the second function call, the same list is reopened and the editing continues, since we want to form the union of bad volumes across the AP-PA pair of dsets. When all is said and done here, the main thing we will want from this is the string selector of the list of good volumes to keep, which will be called dwi_sel_both_goods.txt:
# I/O path, same as before
set path_P_ss = data_proc/SUBJ_001
fat_proc_select_vols \
-in_dwi $path_P_ss/dwi_00/ap.nii.gz \
-in_img $path_P_ss/dwi_00/ap_sepscl.sag.png \
-prefix $path_P_ss/dwi_01/dwi_sel_ap
fat_proc_select_vols \
-in_dwi $path_P_ss/dwi_00/pa.nii.gz \
-in_img $path_P_ss/dwi_00/pa_sepscl.sag.png \
-in_bads $path_P_ss/dwi_01/dwi_sel_ap_bads.txt \
-prefix $path_P_ss/dwi_01/dwi_sel_both
... and during the running, the following actions were taken in this example (again, this is just an example of applying selection in the GUI):
After the first function has started and some clicks have been made (described in caption):
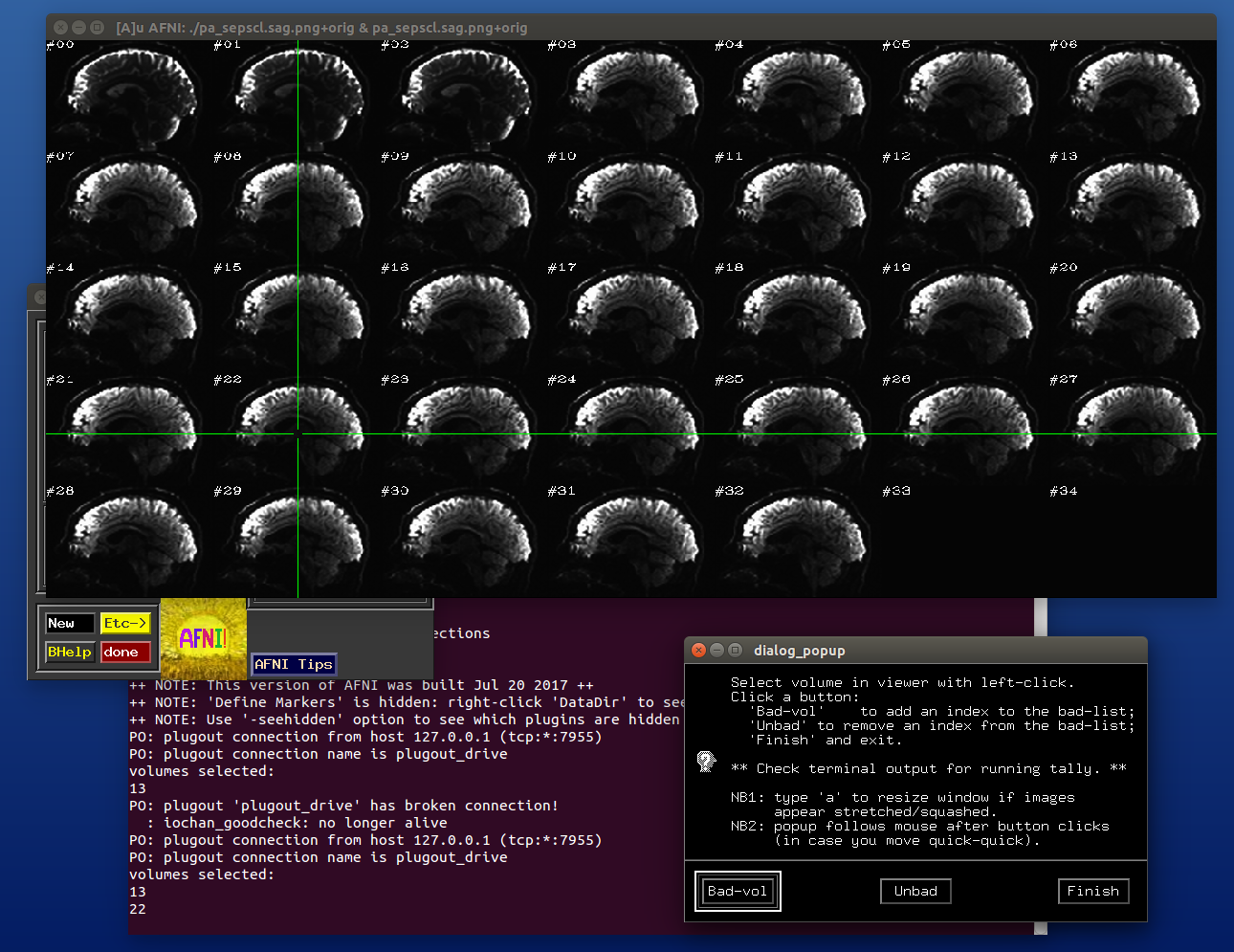
After the first function has finished and the second function has just executed– no clicks made yet:
After some clicks have been made during the execution of the second function:
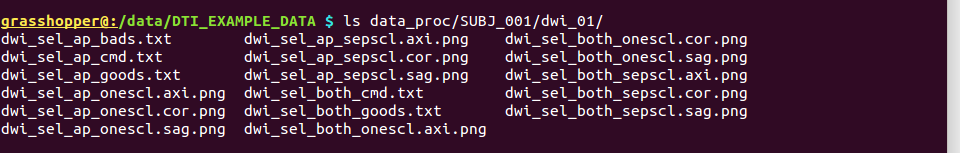
-> produces one new directory in ‘data_proc/SUBJ_001/’, called “dwi_01/”:
Directory structure for example data set |
|---|
|
Output files made by calls to fat_proc_select_vols commands for both the AP and PA data. |
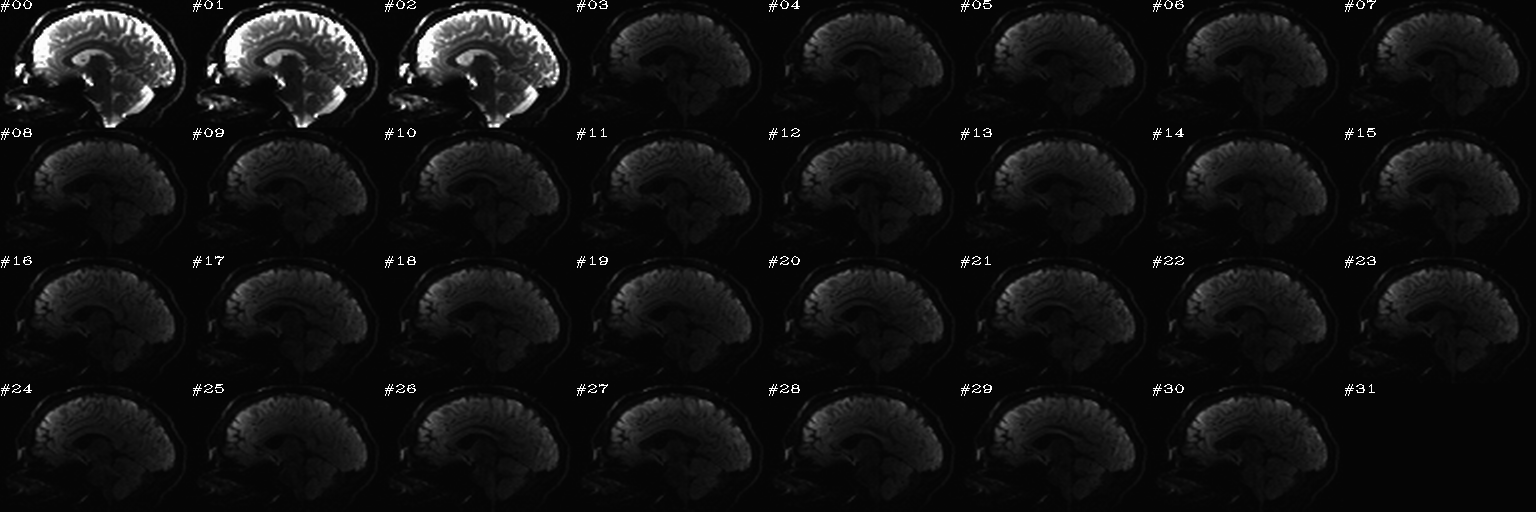
It contains the following outputs for the AP data, and analogous outputs for the PA (=”both”) dsets, but we also note that the “dwi_sel_both_goods.txt” file contains the complement of the union of ‘bad’ selections from both the AP and PA selection, and therefore the the PA (=”both”) images have fewer volumes here.
fat_proc_filter_dwis: apply selection filter to keep goodies¶
Once the string of “good” values to keep in the data set has been made and stored in a simple text file, it can be applied to both a 4D DWI file and some form of the gradient information. For the latter, here we choose to use the TORTOISE-style b-matrix, which contains both the gradient and DW b-value information, because we aim to use TORTOISE’s DIFFPREP in the subsequent step of DWI processing.
Proc: the filter function will be applied to each of the AP and PA dsets individually, though using the same “selection string” in both cases. Note that the input volumes and b-matrices are in the “data_proc/SUBJ_001/dwi_00/” directory, while the selection string is in the “data_proc/SUBJ_001/dwi_01/” directory:
# I/O path, same as before
set path_P_ss = data_proc/SUBJ_001
# the string of *good* volumes after selecting *bads*
set selstr = `cat $path_P_ss/dwi_01/dwi_sel_both_goods.txt`
# filter from both AP and PA dwi sets, both vols and b-matrices
fat_proc_filter_dwis \
-in_dwi $path_P_ss/dwi_00/ap.nii.gz \
-in_col_matT $path_P_ss/dwi_00/ap_matT.dat \
-select "$selstr" \
-prefix $path_P_ss/dwi_02/ap
fat_proc_filter_dwis \
-in_dwi $path_P_ss/dwi_00/pa.nii.gz \
-in_col_matT $path_P_ss/dwi_00/pa_matT.dat \
-select "$selstr" \
-prefix $path_P_ss/dwi_02/pa
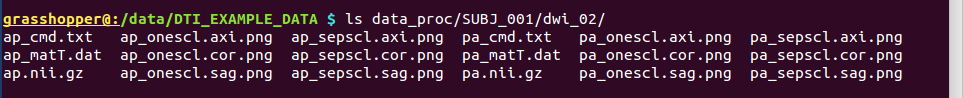
-> produces one new directory in ‘data_proc/SUBJ_001/’, called “dwi_02/”:
Directory structure for example data set |
|---|
|
Output files made by calls to fat_proc_filter_dwis commands for both the AP and PA data. |
It contains the following outputs for the AP data (and analogous outputs for the PA sets):